Molecular characteristics of fibroblast growth factor-fibroblast growth factor receptor-heparin-like glycosaminoglycan complex
- PMID: 10097093
- PMCID: PMC22350
- DOI: 10.1073/pnas.96.7.3658
Molecular characteristics of fibroblast growth factor-fibroblast growth factor receptor-heparin-like glycosaminoglycan complex
Abstract
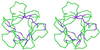
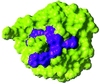
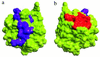
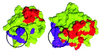
Fibroblast growth factor (FGF) family plays key roles in development, wound healing, and angiogenesis. Understanding of the molecular nature of interactions of FGFs with their receptors (FGFRs) has been seriously limited by the absence of structural information on FGFR or FGF-FGFR complex. In this study, based on an exhaustive analysis of the primary sequences of the FGF family, we determined that the residues that constitute the primary receptor-binding site of FGF-2 are conserved throughout the FGF family, whereas those of the secondary receptor binding site of FGF-2 are not. We propose that the FGF-FGFR interaction mediated by the 'conserved' primary site interactions is likely to be similar if not identical for the entire FGF family, whereas the 'variable' secondary sites, on both FGF as well as FGFR mediates specificity of a given FGF to a given FGFR isoform. Furthermore, as the pro-inflammatory cytokine interleukin 1 (IL-1) and FGF-2 share the same structural scaffold, we find that the spatial orientation of the primary receptor-binding site of FGF-2 coincides structurally with the IL-1beta receptor-binding site when the two molecules are superimposed. The structural similarities between the IL-1 and the FGF system provided a framework to elucidate molecular principles of FGF-FGFR interactions. In the FGF-FGFR model proposed here, the two domains of a single FGFR wrap around a single FGF-2 molecule such that one domain of FGFR binds to the primary receptor-binding site of the FGF molecule, while the second domain of the same FGFR binds to the secondary receptor-binding site of the same FGF molecule. Finally, the proposed model is able to accommodate not only heparin-like glycosaminoglycan (HLGAG) interactions with FGF and FGFR but also FGF dimerization or oligomerization mediated by HLGAG.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

