Horizontal gene transfer among genomes: the complexity hypothesis
- PMID: 10097118
- PMCID: PMC22375
- DOI: 10.1073/pnas.96.7.3801
Horizontal gene transfer among genomes: the complexity hypothesis
Abstract
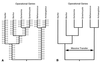
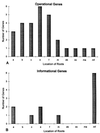
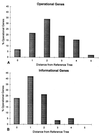
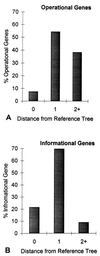
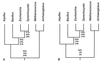
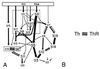
Increasingly, studies of genes and genomes are indicating that considerable horizontal transfer has occurred between prokaryotes. Extensive horizontal transfer has occurred for operational genes (those involved in housekeeping), whereas informational genes (those involved in transcription, translation, and related processes) are seldomly horizontally transferred. Through phylogenetic analysis of six complete prokaryotic genomes and the identification of 312 sets of orthologous genes present in all six genomes, we tested two theories describing the temporal flow of horizontal transfer. We show that operational genes have been horizontally transferred continuously since the divergence of the prokaryotes, rather than having been exchanged in one, or a few, massive events that occurred early in the evolution of prokaryotes. In agreement with earlier studies, we found that differences in rates of evolution between operational and informational genes are minimal, suggesting that factors other than rate of evolution are responsible for the observed differences in horizontal transfer. We propose that a major factor in the more frequent horizontal transfer of operational genes is that informational genes are typically members of large, complex systems, whereas operational genes are not, thereby making horizontal transfer of informational gene products less probable (the complexity hypothesis).
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

