Genomic evolution during a 10,000-generation experiment with bacteria
- PMID: 10097119
- PMCID: PMC22376
- DOI: 10.1073/pnas.96.7.3807
Genomic evolution during a 10,000-generation experiment with bacteria
Abstract
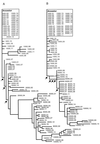
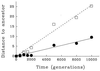
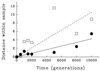
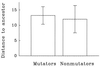
Molecular methods are used widely to measure genetic diversity within populations and determine relationships among species. However, it is difficult to observe genomic evolution in action because these dynamics are too slow in most organisms. To overcome this limitation, we sampled genomes from populations of Escherichia coli evolving in the laboratory for 10,000 generations. We analyzed the genomes for restriction fragment length polymorphisms (RFLP) using seven insertion sequences (IS) as probes; most polymorphisms detected by this approach reflect rearrangements (including transpositions) rather than point mutations. The evolving genomes became increasingly different from their ancestor over time. Moreover, tremendous diversity accumulated within each population, such that almost every individual had a different genetic fingerprint after 10,000 generations. As has been often suggested, but not previously shown by experiment, the rates of phenotypic and genomic change were discordant, both across replicate populations and over time within a population. Certain pivotal mutations were shared by all descendants in a population, and these are candidates for beneficial mutations, which are rare and difficult to find. More generally, these data show that the genome is highly dynamic even over a time scale that is, from an evolutionary perspective, very brief.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

