The Ets transcription factor ERM is Th1-specific and induced by IL-12 through a Stat4-dependent pathway
- PMID: 10097133
- PMCID: PMC22390
- DOI: 10.1073/pnas.96.7.3888
The Ets transcription factor ERM is Th1-specific and induced by IL-12 through a Stat4-dependent pathway
Abstract
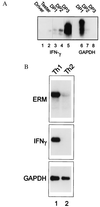
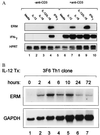
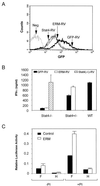
Interleukin 12 (IL-12)-induced T helper 1 (Th1) development requires Stat4 activation. However, antigen-activated Th1 cells can produce interferon gamma (IFN-gamma) independently of IL-12 and Stat4 activation. Thus, in differentiated Th1 cells, factors regulated by IL-12 and Stat4 may be involved in IFN-gamma production. Using subtractive cloning, we identified ERM, an Ets transcription factor, to be a Th1-specific, IL-12-induced gene. IL-12-induction of ERM occurred in wild-type and Stat1-deficient, but not Stat4-deficient, T cells, suggesting ERM is Stat4-inducible. Retroviral expression of ERM did not restore IFN-gamma production in Stat4-deficient T cells, but augmented IFN-gamma expression in Stat4-heterozygous T cells. Ets factors frequently regulate transcription via cooperative interactions with other transcription factors, and ERM has been reported to cooperate with c-Jun. However, in the absence of other transcription factors, ERM augmented expression of an IFN-gamma reporter by only 2-fold. Thus, determining the requirement for ERM in Th1 development likely will require gene targeting.
Figures


References
-
- Abbas A K, Murphy K M, Sher A. Nature (London) 1996;383:787–793. - PubMed
-
- Seder R A, Paul W E. Annu Rev Immunol. 1994;12:635–673. - PubMed
-
- Hsieh C S, Macatonia S E, O’Garra A, Murphy K M. Int Immunol. 1993;5:371–382. - PubMed
-
- Macatonia S E, Hosken N A, Litton M, Vieira P, Hsieh C S, Culpepper J A, Wysocka M, Trinchieri G, Murphy K M, O’Garra A. J Immunol. 1995;154:5071–5079. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

