Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease
- PMID: 10097139
- PMCID: PMC22396
- DOI: 10.1073/pnas.96.7.3922
Constitutive and regulated alpha-secretase cleavage of Alzheimer's amyloid precursor protein by a disintegrin metalloprotease
Abstract
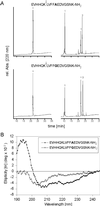
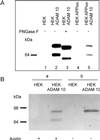
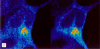
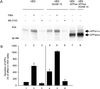
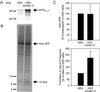
Amyloid beta peptide (Abeta), the principal proteinaceous component of amyloid plaques in brains of Alzheimer's disease patients, is derived by proteolytic cleavage of the amyloid precursor protein (APP). Proteolytic cleavage of APP by a putative alpha-secretase within the Abeta sequence precludes the formation of the amyloidogenic peptides and leads to the release of soluble APPsalpha into the medium. By overexpression of a disintegrin and metalloprotease (ADAM), classified as ADAM 10, in HEK 293 cells, basal and protein kinase C-stimulated alpha-secretase activity was increased severalfold. The proteolytically activated form of ADAM 10 was localized by cell surface biotinylation in the plasma membrane, but the majority of the proenzyme was found in the Golgi. These results support the view that APP is cleaved both at the cell surface and along the secretory pathway. Endogenous alpha-secretase activity was inhibited by a dominant negative form of ADAM 10 with a point mutation in the zinc binding site. Studies with purified ADAM 10 and Abeta fragments confirm the correct alpha-secretase cleavage site and demonstrate a dependence on the substrate's conformation. Our results provide evidence that ADAM 10 has alpha-secretase activity and many properties expected for the proteolytic processing of APP. Increases of its expression and activity might be beneficial for the treatment of Alzheimer's disease.
Figures
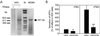
References
-
- Haass C, Selkoe D J. Cell. 1993;75:1039–1042. - PubMed
-
- Selkoe D J. J Biol Chem. 1996;271:18295–18298. - PubMed
-
- Haass C, Koo E H, Mellon A, Hung A Y, Selkoe D J. Nature (London) 1992;357:500–503. - PubMed
-
- Ikezu T, Trapp B D, Song K S, Schlegel A, Lisanti M P, Okamoto T. J Biol Chem. 1998;273:10485–10495. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

