NADH-quinone oxidoreductase: PSST subunit couples electron transfer from iron-sulfur cluster N2 to quinone
- PMID: 10097178
- PMCID: PMC22435
- DOI: 10.1073/pnas.96.7.4149
NADH-quinone oxidoreductase: PSST subunit couples electron transfer from iron-sulfur cluster N2 to quinone
Abstract
The proton-translocating NADH-quinone oxidoreductase (EC 1.6.99.3) is the largest and least understood enzyme complex of the respiratory chain. The mammalian mitochondrial enzyme (also called complex I) contains more than 40 subunits, whereas its structurally simpler bacterial counterpart (NDH-1) in Paracoccus denitrificans and Thermus thermophilus HB-8 consists of 14 subunits. A major unsolved question is the location and mechanism of the terminal electron transfer step from iron-sulfur cluster N2 to quinone. Potent inhibitors acting at this key region are candidate photoaffinity probes to dissect NADH-quinone oxidoreductases. Complex I and NDH-1 are very sensitive to inhibition by a variety of structurally diverse toxicants, including rotenone, piericidin A, bullatacin, and pyridaben. We designed (trifluoromethyl)diazirinyl[3H]pyridaben ([3H]TDP) as our photoaffinity ligand because it combines outstanding inhibitor potency, a suitable photoreactive group, and tritium at high specific activity. Photoaffinity labeling of mitochondrial electron transport particles was specific and saturable. Isolation, protein sequencing, and immunoprecipitation identified the high-affinity specifically labeled 23-kDa subunit as PSST of complex I. Immunoprecipitation of labeled membranes of P. denitrificans and T. thermophilus established photoaffinity labeling of the equivalent bacterial NQO6. Competitive binding and enzyme inhibition studies showed that photoaffinity labeling of the specific high-affinity binding site of PSST is exceptionally sensitive to each of the high-potency inhibitors mentioned above. These findings establish that the homologous PSST of mitochondria and NQO6 of bacteria have a conserved inhibitor-binding site and that this subunit plays a key role in electron transfer by functionally coupling iron-sulfur cluster N2 to quinone.
Figures
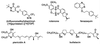

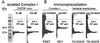
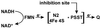
Similar articles
-
Bacterial NADH-quinone oxidoreductases: iron-sulfur clusters and related problems.J Bioenerg Biomembr. 1993 Aug;25(4):347-56. doi: 10.1007/BF00762460. J Bioenerg Biomembr. 1993. PMID: 8226716 Review.
-
Iron-sulfur clusters/semiquinones in complex I.Biochim Biophys Acta. 1998 May 6;1364(2):186-206. doi: 10.1016/s0005-2728(98)00027-9. Biochim Biophys Acta. 1998. PMID: 9593887 Review.
-
Functional coupling of PSST and ND1 subunits in NADH:ubiquinone oxidoreductase established by photoaffinity labeling.Biochim Biophys Acta. 2001 Jul 2;1506(1):79-87. doi: 10.1016/s0005-2728(01)00183-9. Biochim Biophys Acta. 2001. PMID: 11418099
-
Expression and characterization of the 66-kilodalton (NQO3) iron-sulfur subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans.J Biol Chem. 1995 Aug 4;270(31):18264-70. doi: 10.1074/jbc.270.31.18264. J Biol Chem. 1995. PMID: 7629145
-
Expression and characterization of the flavoprotein subcomplex composed of 50-kDa (NQO1) and 25-kDa (NQO2) subunits of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans.J Biol Chem. 1996 Mar 8;271(10):5907-13. doi: 10.1074/jbc.271.10.5907. J Biol Chem. 1996. PMID: 8621464
Cited by
-
Challenges in elucidating structure and mechanism of proton pumping NADH:ubiquinone oxidoreductase (complex I).J Bioenerg Biomembr. 2008 Oct;40(5):475-83. doi: 10.1007/s10863-008-9171-9. Epub 2008 Nov 4. J Bioenerg Biomembr. 2008. PMID: 18982432 Review.
-
Redox-dependent change of nucleotide affinity to the active site of the mammalian complex I.Biochemistry. 2007 Sep 25;46(38):10971-8. doi: 10.1021/bi7009822. Epub 2007 Aug 31. Biochemistry. 2007. PMID: 17760425 Free PMC article.
-
EPR characterization of ubisemiquinones and iron-sulfur cluster N2, central components of the energy coupling in the NADH-ubiquinone oxidoreductase (complex I) in situ.J Bioenerg Biomembr. 2002 Jun;34(3):193-208. doi: 10.1023/a:1016083419979. J Bioenerg Biomembr. 2002. PMID: 12171069
-
Probing the ubiquinone reduction site in bovine mitochondrial complex I using a series of synthetic ubiquinones and inhibitors.J Bioenerg Biomembr. 2001 Jun;33(3):223-31. doi: 10.1023/a:1010735019982. J Bioenerg Biomembr. 2001. PMID: 11695832 Review.
-
Biotoxicity of Cry1Ab protein on wolf spider Pardosa pseudoannulata.Ecotoxicology. 2017 Dec;26(10):1336-1343. doi: 10.1007/s10646-017-1858-4. Epub 2017 Oct 17. Ecotoxicology. 2017. PMID: 29043472
References
-
- Walker J E. Q Rev Biophys. 1992;25:253–324. - PubMed
-
- Ernster, L., Luft, R. & Orrenius, S., eds. (1995) Mitochondrial Diseases, Nobel Symposium 90, Biochim. Biophys. Acta (Special Issue) 1271, 1–292. - PubMed
-
- Howell N. J Bioenerg Biomembr. 1997;29:165–173. - PubMed
-
- Robinson B H. Biochim Biophys Acta. 1998;1364:271–286. - PubMed
-
- Singer T P, Castagnoli N, Jr, Ramsay R R, Trevor A J. J Neurochem. 1987;49:1–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

