Cis-regulatory elements of the mitotic regulator, string/Cdc25
- PMID: 10101114
- PMCID: PMC10176497
- DOI: 10.1242/dev.126.9.1793
Cis-regulatory elements of the mitotic regulator, string/Cdc25
Abstract
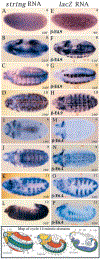
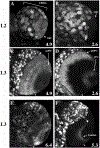
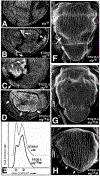
Mitosis in most Drosophila cells is triggered by brief bursts of transcription of string (stg), a Cdc25-type phosphatase that activates the mitotic kinase, Cdk1 (Cdc2). To understand how string transcription is regulated, we analyzed the expression of string-lacZ reporter genes covering approximately 40 kb of the string locus. We also tested protein coding fragments of the string locus of 6 kb to 31.6 kb for their ability to complement loss of string function in embryos and imaginal discs. A plethora of cis-acting elements spread over >30 kb control string transcription in different cells and tissue types. Regulatory elements specific to subsets of epidermal cells, mesoderm, trachea and nurse cells were identified, but the majority of the string locus appears to be devoted to controlling cell proliferation during neurogenesis. Consistent with this, compact promotor-proximal sequences are sufficient for string function during imaginal disc growth, but additional distal elements are required for the development of neural structures in the eye, wing, leg and notum. We suggest that, during evolution, cell-type-specific control elements were acquired by a simple growth-regulated promoter as a means of coordinating cell division with developmental processes, particularly neurogenesis.
Figures



Similar articles
-
Drosophila development pulls the strings of the cell cycle.Bioessays. 1995 Jun;17(6):553-6. doi: 10.1002/bies.950170613. Bioessays. 1995. PMID: 7575498 Review.
-
Zygotic degradation of two maternal Cdc25 mRNAs terminates Drosophila's early cell cycle program.Genes Dev. 1996 Aug 1;10(15):1966-77. doi: 10.1101/gad.10.15.1966. Genes Dev. 1996. PMID: 8756353
-
Tribbles, a cell-cycle brake that coordinates proliferation and morphogenesis during Drosophila gastrulation.Curr Biol. 2000 Jun 1;10(11):623-9. doi: 10.1016/s0960-9822(00)00502-9. Curr Biol. 2000. PMID: 10837248
-
Transcriptional regulation of string (cdc25): a link between developmental programming and the cell cycle.Development. 1994 Nov;120(11):3131-43. doi: 10.1242/dev.120.11.3131. Development. 1994. PMID: 7720557 Free PMC article.
-
Pattern- and growth-linked cell cycles in Drosophila development.Novartis Found Symp. 2001;237:3-12; discussion 12-8, 36-42. doi: 10.1002/0470846666.ch2. Novartis Found Symp. 2001. PMID: 11444048 Review.
Cited by
-
Developmental regulation of cell type-specific transcription by novel promoter-proximal sequence elements.Genes Dev. 2020 May 1;34(9-10):663-677. doi: 10.1101/gad.335331.119. Epub 2020 Mar 26. Genes Dev. 2020. PMID: 32217666 Free PMC article.
-
Myc in model organisms: a view from the flyroom.Semin Cancer Biol. 2006 Aug;16(4):303-12. doi: 10.1016/j.semcancer.2006.07.010. Epub 2006 Jul 14. Semin Cancer Biol. 2006. PMID: 16916612 Free PMC article. Review.
-
Robustness of cell cycle control and flexible orders of signaling events.Sci Rep. 2015 Sep 30;5:14627. doi: 10.1038/srep14627. Sci Rep. 2015. PMID: 26419873 Free PMC article.
-
Notch-dependent downregulation of the homeodomain gene cut is required for the mitotic cycle/endocycle switch and cell differentiation in Drosophila follicle cells.Development. 2005 Oct;132(19):4299-308. doi: 10.1242/dev.02015. Epub 2005 Sep 1. Development. 2005. PMID: 16141223 Free PMC article.
-
Pleiotropy of the Drosophila melanogaster foraging gene on larval feeding-related traits.J Neurogenet. 2018 Sep;32(3):256-266. doi: 10.1080/01677063.2018.1500572. Epub 2018 Oct 10. J Neurogenet. 2018. PMID: 30303018 Free PMC article.
References
-
- Arora K and Nüsslein-Volhard C (1992). Altered mitotic domains reveal fate map changes in Drosophila embryos mutant for zygotic dorsoventral patterning genes. Development 114, 1003–1024. - PubMed
-
- Bodmer R, Carretto R and Jan YN (1989). Neurogenesis of the peripheral nervous system in Drosophila embryos: DNA replication patterns and cell lineages. Neuron 3, 21–32. - PubMed
-
- Breeden L (1996). Start-specific transcription in yeast. Curr. Topics Microbiol. Immunol 208, 95–127. - PubMed
-
- Campos-Ortega JA and Hartenstein V (1985). The Embryonic Development of Drosophila melanogaster Berlin: Springer-Verlag.
-
- Crozatier M, Valle D, Dubois L, Ibnsouda S and Vincent A (1996). collier, a novel regulator of Drosophila head development, is expressed in a single mitotic domain. Curr. Biol 6, 707–718. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

