Complexity in biological signaling systems
- PMID: 10102825
- PMCID: PMC3773983
- DOI: 10.1126/science.284.5411.92
Complexity in biological signaling systems
Abstract
Biological signaling pathways interact with one another to form complex networks. Complexity arises from the large number of components, many with isoforms that have partially overlapping functions; from the connections among components; and from the spatial relationship between components. The origins of the complex behavior of signaling networks and analytical approaches to deal with the emergent complexity are discussed here.
Figures

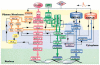
References
-
- Alex LA, Simon MI. Trends Genet. 1994;10:133. - PubMed
-
- Segel IH. Enzyme Kinetics–Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. Wiley; New York: 1975.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

