Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104
- PMID: 10103189
- PMCID: PMC89215
- DOI: 10.1128/AAC.43.4.846
Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104
Abstract
Salmonella typhimurium phage type DT104 has become an important emerging pathogen. Isolates of this phage type often possess resistance to ampicillin, chloramphenicol, streptomycin, sulfonamides, and tetracycline (ACSSuT resistance). The mechanism by which DT104 has accumulated resistance genes is of interest, since these genes interfere with treatment of DT104 infections and might be horizontally transferred to other bacteria, even to unrelated organisms. Previously, several laboratories have shown that the antibiotic resistance genes of DT104 are chromosomally encoded and involve integrons. The antibiotic resistance genes conferring the ACSSuT-resistant phenotype have been cloned and sequenced. These genes are grouped within two district integrons and intervening plasmid-derived sequences. This sequence is potentially useful for detection of multiresistant DT104.
Figures
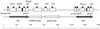

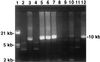
References
-
- Angulo F. Multidrug-resistant Salmonella typhimurium definitive type 104. Emerg Infect Dis. 1997;3:414.
-
- Ferber D. New hunt for the roots of resistance. Science. 1998;280:27. - PubMed
-
- Glynn M K, Bopp C, Dewitt W, Dabney P, Molktar M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. - PubMed
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

