Selection of clc, cba, and fcb chlorobenzoate-catabolic genotypes from groundwater and surface waters adjacent to the Hyde park, Niagara Falls, chemical landfill
- PMID: 10103260
- PMCID: PMC91230
- DOI: 10.1128/AEM.65.4.1627-1635.1999
Selection of clc, cba, and fcb chlorobenzoate-catabolic genotypes from groundwater and surface waters adjacent to the Hyde park, Niagara Falls, chemical landfill
Abstract
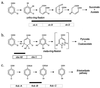
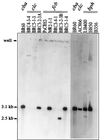
The frequency of isolation of three nonhomologous chlorobenzoate catabolic genotypes (clc, cba, and fcb) was determined for 464 isolates from freshwater sediments and groundwater in the vicinity of the Hyde Park industrial landfill site in the Niagara watershed. Samples were collected from both contaminated and noncontaminated sites during spring, summer, and fall and enriched at 4, 22, or 32 degrees C with micromolar to millimolar concentrations of chlorobenzoates and 3-chlorobiphenyl (M. C. Peel and R. C. Wyndham, Microb. Ecol: 33:59-68, 1997). Hybridization at moderate stringency to restriction-digested genomic DNA with DNA probes revealed the chlorocatechol 1,2-dioxygenase operon (clcABD), the 3-chlorobenzoate 3,4-(4,5)-dioxygenase operon (cbaABC), and the 4-chlorobenzoate dehalogenase (fcbB) gene in isolates enriched from all contaminated sites in the vicinity of the industrial landfill. Nevertheless, the known genes were found in less than 10% of the isolates from the contaminated sites, indicating a high level of genetic diversity in the microbial community. The known genotypes were not enriched from the noncontaminated control sites nearby. The clc, cba, and fcb isolates were distributed across five phenotypically distinct groups based on Biolog carbon source utilization, with the breadth of the host range decreasing in the order clc > cba > fcb. Restriction fragment length polymorphism (RFLP) patterns showed that the cba genes were conserved in all isolates whereas the clc and fcb genes exhibited variation in RFLP patterns. These observations are consistent with the recent spread of the cba genes by horizontal transfer as part of transposon Tn5271 in response to contaminant exposure at Hyde Park. Consistent with this hypothesis, IS1071, the flanking element in Tn5271, was found in all isolates that carried the cba genes. Interestingly, IS1071 was also found in a high proportion of isolates from Hyde Park carrying the clc and fcb genes, as well as in type strains carrying the clcABD operon and the biphenyl (bph) catabolic genes.
Figures

References
-
- Assinder S J, Williams P A. The TOL plasmids: determinants of the catabolism of toluene and xylenes. Adv Microb Physiol. 1990;31:1–69. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989.
-
- Babbitt, P. C., G. L. Kenyon, B. M. Martin, H. Charest, M. Sylvestre, J. D. Scholten, K.-H. Chang, P.-H. Liang, and D. Dunaway-Mariano. 1993. Genpept accession number 419530 (PIR: locus B42560).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

