Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures
- PMID: 10103273
- PMCID: PMC91243
- DOI: 10.1128/AEM.65.4.1721-1730.1999
Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures
Abstract
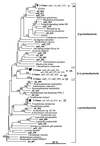
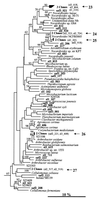
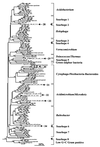
Bacterial community structure and diversity in rhizospheres in two types of grassland, distinguished by both plant species and fertilization regimen, were assessed by performing a 16S ribosomal DNA (rDNA) sequence analysis of DNAs extracted from triplicate soil plots. PCR products were cloned, and 45 to 48 clones from each of the six libraries were partially sequenced. Phylogenetic analysis of the resultant 275 clone sequences indicated that there was considerable variation in abundance in replicate unfertilized, unimproved soil samples and fertilized, improved soil samples but that there were no significant differences in the abundance of any phylogenetic group. Several clone sequences were identical in the 16S rDNA region analyzed, and the clones comprised eight pairs of duplicate clones and two sets of triplicate clones. Many clones were found to be most closely related to environmental clones obtained in other studies, although three clones were found to be identical to culturable species in databases. The clones were clustered into operational taxonomic units at a level of sequence similarity of >97% in order to quantify diversity. In all, 34 clusters containing two or more sequences were identified, and the largest group contained nine clones. A number of diversity, dominance, and evenness indices were calculated, and they all indicated that diversity was high, reflecting the low coverage of rDNA libraries achieved. Differences in diversity between sample types were not observed. Collector's curves, however, indicated that there were differences in the underlying community structures; in particular, there was reduced diversity of organisms of the alpha subdivision of the class Proteobacteria (alpha-proteobacteria) in improved soils.
Figures


References
-
- Atlas R M, Horowitz A, Krichevsky M, Bej A K. Response of microbial populations to environmental disturbance. Microb Ecol. 1991;22:249–256. - PubMed
-
- Brewer A, Williamson M. A new relationship for rarefaction. Biodiv Conserv. 1994;3:373–379.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

