Evidence for a relatively random array of human chromosomes on the mitotic ring
- PMID: 10189364
- PMCID: PMC2148215
- DOI: 10.1083/jcb.145.1.1
Evidence for a relatively random array of human chromosomes on the mitotic ring
Abstract
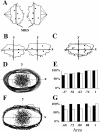
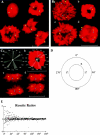
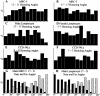
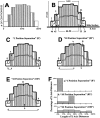
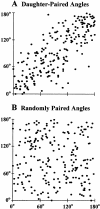
We used fluorescence in situ hybridization (FISH) to study the positions of human chromosomes on the mitotic rings of cultured human lymphocytes, MRC-5 fibroblasts, and CCD-34Lu fibroblasts. The homologous chromosomes of all three cell types had relatively random positions with respect to each other on the mitotic rings of prometaphase rosettes and anaphase cells. Also, the positions of the X and Y chromosomes, colocalized with the somatic homologues in male cells, were highly variable from one mitotic ring to another. Although random chromosomal positions were found in different pairs of CCD-34Lu and MRC-5 late-anaphases, the separations between the same homologous chromosomes in paired late-anaphase and telophase chromosomal masses were highly correlated. Thus, although some loose spatial associations of chromosomes secondary to interphase positioning may exist on the mitotic rings of some cells, a fixed order of human chromosomes and/or a rigorous separation of homologous chromosomes on the mitotic ring are not necessary for normal mitosis. Furthermore, the relative chromosomal positions on each individual metaphase plate are most likely carried through anaphase into telophase.
Figures


References
-
- Bajer A, Molè-Bajer J. Cine-micrographic studies of mitosis in endosperm. II. Chromosome cytoplasmic and Brownian movements. Chromosoma. 1956;7:558–607.
-
- Bajer, A.S., and J. Molè-Bajer. 1981. Mitoses: studies of living cells: a revision of basic concepts. In Mitoses/Cytokineses. A.M. Zimmerman and A. Forer, editors, Academic Press, New York. 227–299.
-
- Barr ML, Bertram EG. A morphological distinction between neurons of the male and female, and the behavior of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676–677. - PubMed
-
- Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science. 1988;242:1687–1691. - PubMed
-
- Boveri T. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch Zellforschung. 1909;3:181–268.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

