Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus
- PMID: 10196248
- PMCID: PMC104131
- DOI: 10.1128/JVI.73.5.3582-3586.1999
Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus
Abstract
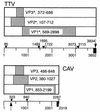
The sequence data (H. Okamoto et al., Hepatol. Res. 10:1-16, 1998) of a newly discovered single-stranded DNA virus, TT virus (TTV), showed that it did not have the terminal structure typical of a parvovirus. Elucidation of the complete genome structure was necessary to understand the nature of TTV. We obtained a 1.0-kb amplified product from serum samples of four TTV carriers by an inverted, nested long PCR targeted for nucleotides (nt) 3025 to 3739 and 1 to 216 of TTV. The sequence of a clone obtained from serum sample TA278 was compared with those registered in GenBank. The complete circular TTV genome contained a novel sequence of 113 nt (nt 3740 to 3852 [=0]) in between the known 3'- and 5'-end arms, forming a 117-nt GC-rich stretch (GC content, 90.6% at nt 3736 to 3852). We found a 36-nt stretch (nt 3816 to 3851) with an 80.6% similarity to chicken anemia virus (CAV) (nt 2237 to 2272 of M55918), a vertebrate circovirus. A putative SP-1 site was located at nt 3834 to 3839, followed by a TATA box at nt 85 to 90, the first initiation codon of a putative VP2 at nt 107 to 109, the termination codon of a putative VP1 at nt 2899 to 2901, and a poly(A) signal at nt 3073 to 3078. The arrangement was similar to that of CAV. Furthermore, several AP-2 and ATF/CREB binding sites and an NF-kappaB site were arranged around the GC-rich region in both TTV and CAV. The data suggested that TTV is circular and similar to CAV in its genomic organization, implying that TTV is the first human circovirus.
Figures




References
-
- Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2173–2197.
-
- Charlton M, Adjei P, Poterucha J, Zein N, Moore B, Therneau T, Krom R, Wiesner R. TT-virus infection in North American blood donors, patients with fulminant hepatic failure, and cryptogenic cirrhosis. Hepatology. 1998;28:839–842. - PubMed
-
- Claessens J A, Schrier C C, Mockett A P, Jagt E H, Sondermeijer P J. Molecular cloning and sequence analysis of the genome of chicken anaemia agent. J Gen Virol. 1991;72:2003–2006. - PubMed
-
- Cossart Y. TTV a common virus, but pathogenic? Lancet. 1998;352:164. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

