Transduction of human progenitor hematopoietic stem cells by human immunodeficiency virus type 1-based vectors is cell cycle dependent
- PMID: 10196257
- PMCID: PMC104140
- DOI: 10.1128/JVI.73.5.3649-3660.1999
Transduction of human progenitor hematopoietic stem cells by human immunodeficiency virus type 1-based vectors is cell cycle dependent
Abstract
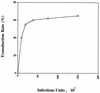
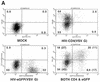
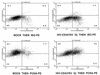
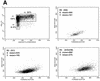
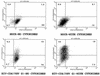
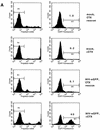
Human immunodeficiency virus (HIV) type 1 vectors are highly efficient in their ability to transduce human progenitor hematopoietic stem cells (PHSC). Although mitosis was not required for transduction of these cells, transduction rates were much greater once cells had been cultured in the presence of cytokines. Transduction rates, however, rarely exceeded 70%. We demonstrate here that there is a distinct subpopulation that is more easily transduced by HIV vectors. These cells were distinguished by a disproportionate population in the S/G2/M phases of the cell cycle. By sorting them prior to transduction, we found that those cells in either the G1 or S/G2/M fraction were more readily transduced than G0 cells. Maintaining the cells in G0 by omitting cytokines from the medium reduced transduction rates by up to 10-fold. Addition of cytokines to the medium immediately after transduction did not improve the transduction efficiency as measured by expression of the transgene. Analysis of replication intermediates indicated that the block to transduction of G0 cells operated near the time of initiation of reverse transcription. These results suggest that although lentivirus vectors can transduce nondividing PHSC, transduction efficiency is severalfold greater once the cells exit G0 and enter G1. Further characterization of these more transducible cells and identification of the cellular factors responsible may enhance transduction while maintaining the pluripotentiality of the PHSC.
Figures









References
-
- Barankiewicz J, Cohen A. Purine nucleotide metabolism in phytohemagglutin-induced human T lymphocytes. Arch Biochem Biophys. 1987;258:167–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

