The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport
- PMID: 10196305
- PMCID: PMC104188
- DOI: 10.1128/JVI.73.5.4090-4100.1999
The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport
Abstract
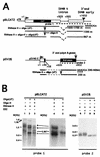
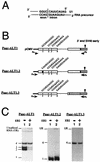
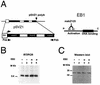
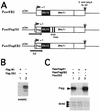
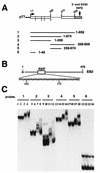
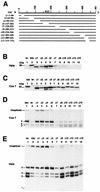
The Epstein-Barr virus BMLF1 gene product EB2 has been shown to efficiently transform immortalized Rat1 and NIH 3T3 cells, to bind RNA, and to shuttle from the nucleus to the cytoplasm. In transient-expression assays EB2 seems to affect mRNA nuclear export of intronless RNAs and pre-mRNA 3' processing, but no direct proof of EB2 being involved in RNA processing and transport has been provided, and no specific functional domain of EB2 has been mapped. Here we significantly extend these findings and directly demonstrate that (i) EB2 inhibits the cytoplasmic accumulation of mRNAs, but only if they are generated from precursors containing weak (cryptic) 5' splice sites, (ii) EB2 has no effect on the cytoplasmic accumulation of mRNA generated from precursors containing constitutive splice sites, and (iii) EB2 has no effect on the 3' processing of precursor RNAs containing canonical and noncanonical cleavage-polyadenylation signals. We also show that in the presence of EB2, intron-containing and intronless RNAs accumulate in the cytoplasm. EB2 contains an Arg-X-Pro tripeptide repeated eight times, similar to that described as an RNA-binding domain in the herpes simplex virus type 1 protein US11. As glutathione S-transferase fusion proteins, both EB2 and the Arg-X-Pro repeat bound RNA in vitro. However, by using EB2 deletion mutants, we demonstrated that the effect of EB2 on splicing and RNA transport requires the C-terminal half of the protein but not the Arg-X-Pro repeat.
Figures



References
-
- Baer R, Bankier A, Biggin M, Deininger P, Farrell P, Gibson T, Hatfull G, Hudson G, Satchwell S, Deguin C, Tuffnell P, Barrell B. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. - PubMed
-
- Boshart M, Klüppel M, Schmidt A, Schütz G, Lucckow B. Reporter constructs with low background activity utilizing the cat gene. Gene. 1992;110:129–130. - PubMed
-
- Boss H, Berger R, Kuklik R C, Ifner T, Mueller L N. Enhancement of Epstein-Barr virus membrane protein (LMP) expression by serum, TPA, or n-butyrate in latently infected cells. Virology. 1987;159:161–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

