CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters
- PMID: 10196318
- PMCID: PMC104201
- DOI: 10.1128/JVI.73.5.4220-4229.1999
CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters
Abstract
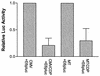
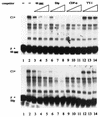
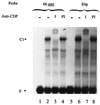
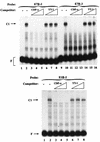
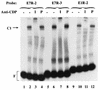
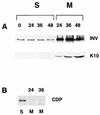
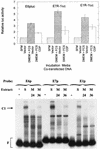
Expression of human papillomavirus genes increases as the target cell, the keratinocyte, differentiates. CCAAT displacement protein (CDP) is a cellular protein which has been shown in other cell types to negatively regulate gene expression in undifferentiated cells but not in differentiated cells. We have previously shown that a 66-bp purine-thymidine-rich sequence (the 66-mer) binds CDP and negatively regulates the human papillomavirus type 6 (HPV-6) E6 promoter (S. Pattison, D. G. Skalnik, and A. Roman, J. Virol. 71:2013-2022, 1997). Cotransfection experiments with a plasmid expressing luciferase from the HPV-6 E6, E7, or E1 regulatory region and a plasmid carrying the CDP gene indicate that CDP represses transcription from all three HPV-6 promoters. Using electrophoretic mobility shift assays (EMSAs), we have shown that CDP binds HPV-6 both upstream and downstream of the E6, E7, and E1 transcription initiation start sites. Furthermore, when keratinocytes were induced to differentiate, all three promoter activities increased. Consistent with this, immunoblotting and EMSAs revealed that endogenous nucleus CDP and, correspondingly, DNA binding activity decreased when keratinocytes were induced to differentiate. The elevated promoter activities were abrogated by exogenously transfected CDP. Our data demonstrate that CDP fulfills the requirement of a differentiation-dependent negative regulator that could tie the HPV life cycle to keratinocyte differentiation.
Figures





References
-
- Andersen B, Hariri A, Pittelkow M R, Rosenfeld M G. Characterization of Skn-1a/i POU domain factors and linkage to papillomavirus gene expression. J Biol Chem. 1997;272:15905–15913. - PubMed
-
- Andres V, Chiara M D, Mahdavi V. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 1994;8:245–257. - PubMed
-
- Andres V, Nadal-Ginard B, Mahdavi V. Clox, a mammalian homeobox gene related to Drosophila cut, encodes DNA-binding regulatory proteins differentially expressed during development. Development. 1992;116:321–334. - PubMed
-
- Apt D, Watts R M, Suske G, Bernard H U. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology. 1996;224:281–291. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

