Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes
- PMID: 10196339
- PMCID: PMC104222
- DOI: 10.1128/JVI.73.5.4413-4426.1999
Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes
Abstract
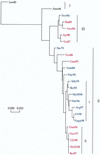
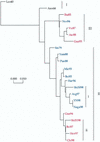
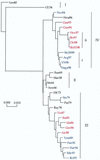
Phylogenetic profiles of the genes coding for the hemagglutinin (HA) protein, nucleoprotein (NP), matrix (M) protein, and nonstructural (NS) proteins of influenza B viruses isolated from 1940 to 1998 were analyzed in a parallel manner in order to understand the evolutionary mechanisms of these viruses. Unlike human influenza A (H3N2) viruses, the evolutionary pathways of all four genes of recent influenza B viruses revealed similar patterns of genetic divergence into two major lineages. Although evolutionary rates of the HA, NP, M, and NS genes of influenza B viruses were estimated to be generally lower than those of human influenza A viruses, genes of influenza B viruses demonstrated complex phylogenetic patterns, indicating alternative mechanisms for generation of virus variability. Topologies of the evolutionary trees of each gene were determined to be quite distinct from one another, showing that these genes were evolving in an independent manner. Furthermore, variable topologies were apparently the result of frequent genetic exchange among cocirculating epidemic viruses. Evolutionary analysis done in the present study provided further evidence for cocirculation of multiple lineages as well as sequestering and reemergence of phylogenetic lineages of the internal genes. In addition, comparison of deduced amino acid sequences revealed a novel amino acid deletion in the HA1 domain of the HA protein of recent isolates from 1998 belonging to the B/Yamagata/16/88-like lineage. It thus became apparent that, despite lower evolutionary rates, influenza B viruses were able to generate genetic diversity among circulating viruses through a combination of evolutionary mechanisms involving cocirculating lineages and genetic reassortment by which new variants with distinct gene constellations emerged.
Figures


References
-
- Betakova T, Nermut M V, Hay A J. The NB protein is an integral component of the membrane of influenza B virus. J Gen Virol. 1996;77:2689–2694. - PubMed
-
- Brassard D L, Leser G P, Lamb R A. Influenza B virus NB glycoprotein is a component of the virion. Virology. 1996;220:350–360. - PubMed
-
- Briedis D J, Lamb R A, Choppin P W. Sequence of RNA segment 7 of the influenza B virus genome: partial amino acid homology between the membrane proteins (M1) of influenza A and B viruses and conservation of a second open reading frame. Virology. 1982;116:581–588. - PubMed
-
- Briedis D J, Tobin M. Influenza B virus genome: complete nucleotide sequence of the influenza B/Lee/40 virus genome RNA segment 5 encoding the nucleoprotein and comparison with the B/Singapore/222/79 nucleoprotein. Virology. 1984;133:448–455. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

