Saturation mutagenesis of the TATA box and upstream activator sequence in the haloarchaeal bop gene promoter
- PMID: 10198017
- PMCID: PMC93679
- DOI: 10.1128/JB.181.8.2513-2518.1999
Saturation mutagenesis of the TATA box and upstream activator sequence in the haloarchaeal bop gene promoter
Abstract
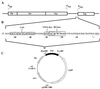
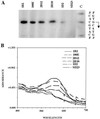
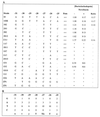
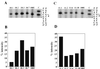
Degenerate oligonucleotides were used to randomize 21 bp of the 53-bp minimal bop promoter in three 7-bp segments, including the putative TATA box and the upstream activator sequence (UAS). The mutagenized bop promoter and the wild-type structural gene and transcriptional terminator were inserted into a shuttle plasmid capable of replication in the halophilic archaeon Halobacterium sp. strain S9. Active promoters were isolated by screening transformants of an orange (Pum- bop) Halobacterium mutant for purple (Pum+ bop+) colonies on agar plates and analyzed for bop mRNA and/or bacteriorhodopsin content. Sequence analysis yielded the consensus sequence 5'-tyT(T/a)Ta-3', corresponding to the promoter TATA box element 30 to 25 bp 5' of the transcription start site. A putative UAS, 5'-ACCcnactagTTnG-3', located 52 to 39 bp 5' of the transcription start site was found to be conserved in active promoters. This study provides direct evidence for the requirement of the TATA box and UAS for bop promoter activity.
Figures

References
-
- Baliga, N. S., and S. DasSarma. Unpublished data.
-
- Baumann P, Qureshi S A, Jackson S P. Transcription: new insights from studies on archaea. Trends Genet. 1995;11:279–283. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

