Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins
- PMID: 10198056
- PMCID: PMC25232
- DOI: 10.1091/mbc.10.4.1043
Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins
Abstract
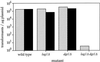
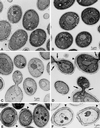
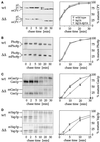
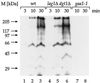
Many eukaryotic cell surface proteins are anchored in the lipid bilayer through glycosylphosphatidylinositol (GPI). GPI anchors are covalently attached in the endoplasmic reticulum (ER). The modified proteins are then transported through the secretory pathway to the cell surface. We have identified two genes in Saccharomyces cerevisiae, LAG1 and a novel gene termed DGT1 (for "delayed GPI-anchored protein transport"), encoding structurally related proteins with multiple membrane-spanning domains. Both proteins are localized to the ER, as demonstrated by immunofluorescence microscopy. Deletion of either gene caused no detectable phenotype, whereas lag1Delta dgt1Delta cells displayed growth defects and a significant delay in ER-to-Golgi transport of GPI-anchored proteins, suggesting that LAG1 and DGT1 encode functionally redundant or overlapping proteins. The rate of GPI anchor attachment was not affected, nor was the transport rate of several non-GPI-anchored proteins. Consistent with a role of Lag1p and Dgt1p in GPI-anchored protein transport, lag1Delta dgt1Delta cells deposit abnormal, multilayered cell walls. Both proteins have significant sequence similarity to TRAM, a mammalian membrane protein thought to be involved in protein translocation across the ER membrane. In vivo translocation studies, however, did not detect any defects in protein translocation in lag1Delta dgt1Delta cells, suggesting that neither yeast gene plays a role in this process. Instead, we propose that Lag1p and Dgt1p facilitate efficient ER-to-Golgi transport of GPI-anchored proteins.
Figures




References
-
- Ash J, Dominguez M, Bergeron JJ, Thomas DY, Bourbonnais Y. The yeast proprotein convertase encoded by YAP3 is a glycophosphatidylinositol-anchored protein that localizes to the plasma membrane. J Biol Chem. 1995;270:20847–20854. - PubMed
-
- Becker DM, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. - PubMed
-
- Belden WJ, Barlowe C. Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J Biol Chem. 1996;271:26939–26946. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

