A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2
- PMID: 10200259
- PMCID: PMC16329
- DOI: 10.1073/pnas.96.8.4313
A MHC-encoded ubiquitin-like protein (FAT10) binds noncovalently to the spindle assembly checkpoint protein MAD2
Abstract
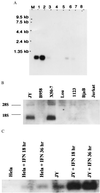
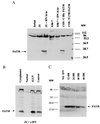
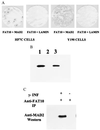
Recently a number of nonclass I genes were discovered in the human MHC class I region. One of these, FAT10, encodes a protein consisting of two domains with homology to ubiquitin. FAT10 mRNA is expressed constitutively in some lymphoblastoid lines and dendritic cells and in certain other cells after gamma-interferon induction. FAT10 protein expression is controlled at several levels including transcription, translation, and protein stability. Yeast two-hybrid screening of a human lymphocyte library and immunoprecipitation studies revealed that FAT10 noncovalently associated with MAD2, a protein implicated in a cell-cycle checkpoint for spindle assembly during anaphase. Thus, FAT10 may modulate cell growth during B cell or dendritic cell development and activation.
Figures


References
-
- Gruen J R, Weissman S M. Blood. 1997;90:4252–4265. - PubMed
-
- Gruen J R, Nalabolu S R, Chu T W, Bowlus C, Fan W, Goei V L, Wei H, Sivakamasundari R, Liu Y-C, Xu H, et al. Genomics. 1996;36:70–85. - PubMed
-
- Fan W, Cai W, Parimoo S, Lennon G G, Weissman S M. Immunogenetics. 1996;44:97–103. - PubMed
-
- Bates E E, Ravel O, Dieu M C, Ho S, Guret C, Bridon J M, Ait-Yahia S, Briere F, Caux C, Banchereau J, et al. Eur J Immunol. 1977;10:2471–2477. - PubMed
-
- Hochstrasser M. Annu Rev Genet. 1997;30:405–439. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

