The NDUFA1 gene product (MWFE protein) is essential for activity of complex I in mammalian mitochondria
- PMID: 10200266
- PMCID: PMC16336
- DOI: 10.1073/pnas.96.8.4354
The NDUFA1 gene product (MWFE protein) is essential for activity of complex I in mammalian mitochondria
Abstract
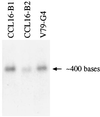
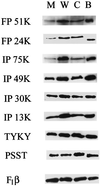
The MWFE polypeptide of mammalian complex I (the proton-translocating NADH-quinone oxidoreductase) is 70 amino acids long, and it is predicted to be a membrane protein. The NDUFA1 gene encoding the MWFE polypeptide is located on the X chromosome. This polypeptide is 1 of approximately 28 "accessory proteins" identified in complex I, which is composed of 42 unlike subunits. It was considered accessory, because it is not one of the 14 polypeptides making up the core complex I; a homologous set of 14 polypeptides can make a fully functional proton-translocating NADH-quinone oxidoreductase in prokaryotes. One MWFE mutant has been identified and isolated from a collection of respiration-deficient Chinese hamster cell mutants. The CCL16-B2 mutant has suffered a deletion that would produce a truncated and abnormal MWFE protein. In these mutant cells, complex I activity is reduced severely (<10%). Complementation with hamster NDUFA1 cDNA restored the rotenone-sensitive complex I activity of these mutant cells to approximately 100% of the parent cell activity. Thus, it is established that the MWFE polypeptide is absolutely essential for an active complex I in mammals.
Figures


References
-
- Yagi T, Yano T, Di Bernardo S, Matsuno-Yagi A. Biochim Biophys Acta. 1998;1364:125–133. - PubMed
-
- Rasmusson A G, Heiser V, Zabaleta E, Brennicke A, Grohmann L. Biochim Biophys Acta. 1998;1364:101–111. - PubMed
-
- Guénebaut V, Schlitt A, Weiss H, Leonard K, Friedrich T. J Mol Biol. 1998;276:105–112. - PubMed
-
- Walker J E. Biochim Biophys Acta. 1995;1271:221–227. - PubMed
-
- Schulte U, Weiss H. Methods Enzymol. 1995;260:3–14. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

