Acyl homoserine-lactone quorum-sensing signal generation
- PMID: 10200267
- PMCID: PMC16337
- DOI: 10.1073/pnas.96.8.4360
Acyl homoserine-lactone quorum-sensing signal generation
Abstract
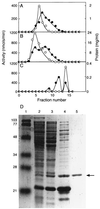
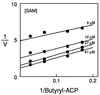
Acyl homoserine lactones (acyl-HSLs) are important intercellular signaling molecules used by many bacteria to monitor their population density in quorum-sensing control of gene expression. These signals are synthesized by members of the LuxI family of proteins. To understand the mechanism of acyl-HSL synthesis we have purified the Pseudomonas aeruginosa RhlI protein and analyzed the kinetics of acyl-HSL synthesis by this enzyme. Purified RhlI catalyzes the synthesis of acyl-HSLs from acyl-acyl carrier proteins and S-adenosylmethionine. An analysis of the patterns of product inhibition indicated that RhlI catalyzes signal synthesis by a sequential, ordered reaction mechanism in which S-adenosylmethionine binds to RhlI as the initial step in the enzymatic mechanism. Because pathogenic bacteria such as P. aeruginosa use acyl-HSL signals to regulate virulence genes, an understanding of the mechanism of signal synthesis and identification of inhibitors of signal synthesis has implications for development of quorum sensing-targeted antivirulence molecules.
Figures



References
-
- Fuqua C, Greenberg E P. Curr Opin Microbiol. 1998;1:183–189. - PubMed
-
- Fuqua C, Winans S C, Greenberg E P. Annu Rev Microbiol. 1996;50:727–751. - PubMed
-
- Sitnikov D M, Schineller J B, Baldwin T O. Mol Microbiol. 1995;17:801–812. - PubMed
-
- Salmond G P C, Bycroft B W, Stewart G S A B, Williams P. Mol Microbiol. 1995;16:615–624. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

