Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology
- PMID: 10200309
- PMCID: PMC16379
- DOI: 10.1073/pnas.96.8.4604
Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology
Abstract
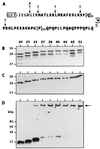
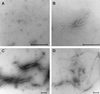
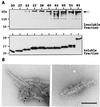
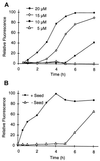
Huntington's disease is a progressive neurodegenerative disorder caused by a polyglutamine [poly(Q)] repeat expansion in the first exon of the huntingtin protein. Previously, we showed that N-terminal huntingtin peptides with poly(Q) tracts in the pathological range (51-122 glutamines), but not with poly(Q) tracts in the normal range (20 and 30 glutamines), form high molecular weight protein aggregates with a fibrillar or ribbon-like morphology, reminiscent of scrapie prion rods and beta-amyloid fibrils in Alzheimer's disease. Here we report that the formation of amyloid-like huntingtin aggregates in vitro not only depends on poly(Q) repeat length but also critically depends on protein concentration and time. Furthermore, the in vitro aggregation of huntingtin can be seeded by preformed fibrils. Together, these results suggest that amyloid fibrillogenesis in Huntington's disease, like in Alzheimer's disease, is a nucleation-dependent polymerization.
Figures

References
-
- Becher M W, Kotzuk J A, Sharp A H, Davies S W, Bates G P, Price D L, Ross C A. Neurobiol Dis. 1997;4:1–11. - PubMed
-
- DiFiglia M, Sapp E, Chase K O, Davies S W, Bates G P, Vonsattel J P, Aronin N. Science. 1997;277:1990–1993. - PubMed
-
- Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Nat Genet. 1998;18:111–117. - PubMed
-
- Li M M, Kobayashi Y, Merry D E, Yamamoto M, Tanaka F, Doyu M, Hashizume Y, Fischbeck K H, Sobue G. Ann Neurol. 1998;44:249–254. - PubMed
-
- Skinner P J, Koshy B T, Cummings C L, Klement I A, Helin K, Servadio A, Zoghbi H Y, Orr H T. Nature (London) 1997;389:971–974. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

