Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals
- PMID: 10200328
- PMCID: PMC16398
- DOI: 10.1073/pnas.96.8.4718
Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals
Abstract
An important effector of Ca2+ signaling in animals and yeast is the Ca2+/calmodulin-dependent protein phosphatase calcineurin. However, the biochemical identity of plant calcineurin remained elusive. Here we report the molecular characterization of AtCBL (Arabidopsis thaliana calcineurin B-like protein) from Arabidopsis. The protein is most similar to mammalian calcineurin B, the regulatory subunit of the phosphatase. AtCBL also shows significant similarity with another Ca2+-binding protein, the neuronal calcium sensor in animals. It contains typical EF-hand motifs with Ca2+-binding capability, as confirmed by in vitro Ca2+-binding assays, and it interacts in vivo with rat calcineurin A in the yeast two-hybrid system. Interaction of AtCBL1 and rat calcineurin A complemented the salt-sensitive phenotype in a yeast calcineurin B mutant. Cloning of cDNAs revealed that AtCBL proteins are encoded by a family of at least six genes in Arabidopsis. Genes for three isoforms were identified in this study. AtCBL1 mRNA was preferentially expressed in stems and roots and its mRNA levels strongly increased in response to specific stress signals such as drought, cold, and wounding. In contrast, AtCBL2 and AtCBL3 are constitutively expressed under all conditions investigated. Our data suggest that AtCBL1 may act as a regulatory subunit of a plant calcineurin-like activity mediating calcium signaling under certain stress conditions.
Figures




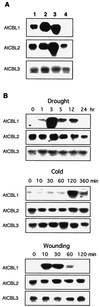
Comment in
-
How plants learn.Proc Natl Acad Sci U S A. 1999 Apr 13;96(8):4216-8. doi: 10.1073/pnas.96.8.4216. Proc Natl Acad Sci U S A. 1999. PMID: 10200239 Free PMC article. Review. No abstract available.
References
-
- Clapham D E. Cell. 1995;80:259–268. - PubMed
-
- Trewavas A, Read N, Campbell A K, Knight M. Biochem Soc Trans. 1996;24:971–974. - PubMed
-
- Hunter T. Cell. 1995;80:225–236. - PubMed
-
- Guerini D. Biochem Biophys Res Commun. 1997;235:271–275. - PubMed
-
- Klee C B, Draetta G F, Hubbard M J. Adv Enzymol. 1988;61:149–200. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

