Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy
- PMID: 10201407
- PMCID: PMC3282055
- DOI: 10.1038/7610
Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy
Abstract
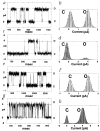
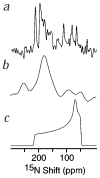
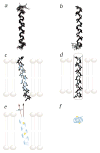
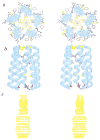
The structures of functional peptides corresponding to the predicted channel-lining M2 segments of the nicotinic acetylcholine receptor (AChR) and of a glutamate receptor of the NMDA subtype (NMDAR) were determined using solution NMR experiments on micelle samples, and solid-state NMR experiments on bilayer samples. Both M2 segments form straight transmembrane alpha-helices with no kinks. The AChR M2 peptide inserts in the lipid bilayer at an angle of 12 degrees relative to the bilayer normal, with a rotation about the helix long axis such that the polar residues face the N-terminal side of the membrane, which is assigned to be intracellular. A model built from these solid-state NMR data, and assuming a symmetric pentameric arrangement of M2 helices, results in a funnel-like architecture for the channel, with the wide opening on the N-terminal intracellular side.
Figures

References
-
- Colquhoun D, Sakmann B. From muscle endplate to brain synapses: a short history of synapses and agonist-activated ion channels. Neuron. 1998;20:381–387. - PubMed
-
- Lena C, Changeux JP. Pathological mutations of nicotinic receptors and nicotine-based therapies for brain disorders. Curr Opin Neurobiol. 1997;7:674–682. - PubMed
-
- Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. - PubMed
-
- Unwin N. Acetylcholine receptor channel imaged in the open state. Nature. 1995;373:37–43. - PubMed
-
- Oiki S, Madison V, Montal M. Bundles of amphipathic transmembrane α-helices as a structural motif for ion-conducting channel proteins: studies on sodium channels and acetylcholine receptors. Proteins. 1990;8:226–236. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
