Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance
- PMID: 10203470
- PMCID: PMC84750
- DOI: 10.1128/JCM.37.5.1274-1279.1999
Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance
Abstract
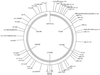
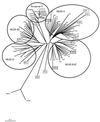
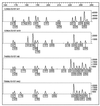
The fluorescent amplified-fragment length polymorphism (FAFLP) assay potentially amplifies a unique set of genome fragments from each bacterial clone. It uses stringently hybridizing primers which carry a fluorescent label. Precise fragment sizing is achieved by the inclusion of an internal size standard in every lane. Therefore, a unique genotype identifier(s) can be found in the form of fragments of precise size or sizes, and these can be generated reproducibly. In order to evaluate the potential of FAFLP as an epidemiological typing method with a valid phylogenetic basis, we applied it to 87 strains of Escherichia coli. These comprised the EcoR collection, which has previously been classified by multilocus enzyme electrophoresis (MLEE) and which represents the genetic diversity of the species E. coli, plus 15 strains of the clinically important serogroup O157. FAFLP with an unlabelled nonselective EcoRI primer (Eco+0) and a labelled selective MseI primer (Mse+TA) gave strain-specific profiles. Fragments of identical sizes (in base pairs) were assumed to be identical, and the genetic distances between the strains were calculated. A phylogenetic tree derived from measure of distance correlated closely with the MLEE groupings of the EcoR collection and placed the verocytotoxin-producing O157 strains on an outlier branch. Our data indicate that FAFLP is suitable for epidemiological investigation of E. coli infection, providing well-defined and reproducible identifiers of genotype for each strain. Since FAFLP objectively samples the whole genome, each strain or isolate can be assigned a place within the broad context of the whole species and can also be subjected to a high-resolution comparison with closely related strains to investigate epidemiological clonality.
Figures
References
-
- Arbeit R, Arthur M, Dunn R, Cheung K, Selander R K, Goldstein R. Resolution of recent divergence among Escherichia coli from related lineages: the application of pulsed field gel electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. - PubMed
-
- Ausubel F M, Brent R, Kingstown R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology I. New York, N.Y: John Wiley & Sons, Inc.; 1989. Preparation of genomic DNA from bacteria.
-
- Cebula T A, Li B, Payne W L, Leclerc L E. ASM Conference on Small Genomes. Washington, D.C: American Society for Microbiology; 1998. Mutators among Escherichia coli and Salmonella enterica: adaptation and emergence of bacterial pathogens; p. 16.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

