Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients
- PMID: 10203499
- PMCID: PMC84792
- DOI: 10.1128/JCM.37.5.1426-1430.1999
Distribution of Porphyromonas gingivalis strains with fimA genotypes in periodontitis patients
Abstract
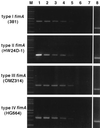
Fimbriae (FimA) of Porphyromonas gingivalis are filamentous components on the cell surface and are thought to play an important role in the colonization and invasion of periodontal tissues. We previously demonstrated that fimA can be classified into four variants (types I to IV) on the basis of the nucleotide sequences of the fimA gene. In the present study, we attempted to detect the four different fimA genes in saliva and plaque samples isolated from patients with periodontitis using the PCR method. Four sets of fimA type-specific primers were designed for the PCR assay. These primers selectively amplified 392-bp (type I), 257-bp (type II), 247-bp (type III), and 251-bp (type IV) DNA fragments of the fimA gene. Positive PCR results were observed with reference strains of P. gingivalis in a type-specific manner. All other laboratory strains of oral and nonoral bacteria gave negative results. The sensitivity of the PCR assay for fimA type-specific detection was between 5 and 50 cells of P. gingivalis. Clinical samples were obtained from saliva and subgingival plaque from deep pockets (>/=4 mm) of 93 patients with periodontitis. Bacterial genomic DNA was isolated from the samples, and the targeted fragments were amplified by PCR. The presence of P. gingivalis was demonstrated in 73 patients (78.5%), and a single fimA gene was detected in most patients. The distribution of the four fimA types among the P. gingivalis-positive patients was as follows: type I, 5.4%; type II, 58.9%; type III, 6. 8%; type IV, 12.3%; types I and II, 6.8%; types II and IV, 2.7%; and untypeable, 6.8%. P. gingivalis with type II fimA was detected more frequently in the deeper pockets, and a significant difference of the occurrence was observed between shallow (4 mm) and deep (>/=8 mm) pockets. These results suggest that P. gingivalis strains that possess type II fimA are significantly more predominant in periodontitis patients, and we speculate that these organisms are involved in the destructive progression of periodontal diseases.
Figures
References
-
- Amano A, Shizukuishi S. Plaque formation: involvement of salivary protein molecules. Dent Outlook. 1996;87:217–227. . (In Japanese.)
-
- Ashimoto A, Chen C, Bakker I, Slots J. Polymerase chain reaction detection of 8 putative periodontal pathogens in subgingival plaque of gingivitis and advanced periodontitis lesions. Oral Microbiol Immunol. 1996;11:266–273. - PubMed
-
- Conrads G, Mutters R, Fischer J, Brauner A, Lutticken R, Lampert F. PCR reaction and dot-blot hybridization to monitor the distribution of oral pathogens within plaque samples of periodontally healthy individuals. J Periodontol. 1996;67:994–1003. - PubMed
-
- Darveau R P, Tanner A, Page R C. The microbial challenge in periodontitis. Periodontol 2000. 1997;14:12–32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

