A highly sensitive assay for detection and quantitation of human cytomegalovirus DNA in serum and plasma by PCR and electrochemiluminescence
- PMID: 10203511
- PMCID: PMC84811
- DOI: 10.1128/JCM.37.5.1489-1497.1999
A highly sensitive assay for detection and quantitation of human cytomegalovirus DNA in serum and plasma by PCR and electrochemiluminescence
Abstract
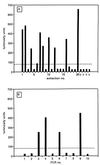
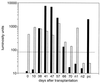
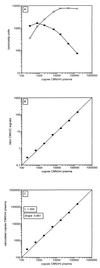
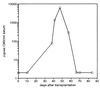
We describe a diagnostic PCR assay (D-PCR) and a quantitative PCR assay (Q-PCR) for the detection of human cytomegalovirus (CMV) in plasma and serum. In the D-PCR, DNA was purified from plasma or serum together with internal control (IC) DNA, which monitored both DNA extraction efficiency and PCR efficiency. DNA was subjected to PCR with a single primer pair, and the amount of PCR products was determined by electrochemiluminescence (ECL) in the QPCR System 5000 (Perkin-Elmer) after hybridization with Tris (2,2'-bipyridine) ruthenium (II) chelate-labeled probes. The lower limit of sensitivity of the D-PCR was reached at about 25 CMV particles/ml. Even with extremely low DNA inputs (four molecules of IC DNA/200 microl of plasma), very high yields (near 100%) were reached. DNA extracted from specimens that were CMV positive by the D-PCR was subsequently used in the Q-PCR, which was similar to the D-PCR. The viral load was calculated directly from the ratio of CMV and IC signals obtained by ECL. The Q-PCR assay is quantitative in the range of 100 to 150,000 copies of CMV/ml, independent of the anticoagulant. Interassay variation, intra-assay variation, and interspecimen variation were about 25%, suggesting that the Q-PCR will reliably detect fourfold differences in viral load. Comparison of paired serum and plasma specimens from CMV-infected individuals showed that serum CMV loads were frequently more than 10-fold lower than plasma CMV loads.
Figures


References
-
- Akrigg A, Wilkinson G W G, Oram J D. The structure of the major immediate early gene of human cytomegalovirus strain AD169. Virus Res. 1985;2:107–121. - PubMed
-
- Aspin M A, Gallez-Hawkins G M, Giugni T D, Tegtmeier B, Lang D J, Schmidt G M, Forman S J, Zaia J. Comparison of plasma PCR and bronchoalveolar lavage fluid culture for detection of cytomegalovirus infection in adult bone marrow transplant recipients. J Clin Microbiol. 1994;32:2266–2269. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

