The linking regions of EBNA1 are essential for its support of replication and transcription
- PMID: 10207059
- PMCID: PMC84128
- DOI: 10.1128/MCB.19.5.3349
The linking regions of EBNA1 are essential for its support of replication and transcription
Abstract
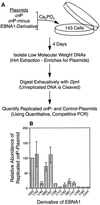
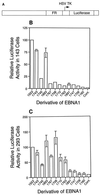
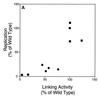
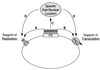
The ability of distant cis-acting DNA elements to interact functionally has been proposed to be mediated by the interaction of proteins associated site specifically with those cis-acting elements. We have found that the DNA-linking regions of EBNA1 are essential for its contribution to both replication and transcription. The synthesis of plasmids containing the Epstein-Barr virus (EBV) origin of plasmid replication (oriP) can be mediated entirely by the cellular machinery; however, the replicated molecules are lost rapidly from proliferating cells. When EBNA1 is provided in trans, plasmids containing oriP (oriP plasmids) are synthesized during repeated S phases, and the newly formed daughter molecules are precisely segregated to the daughter cells. The contribution(s) of EBNA1 to the stable replication of oriP plasmids is therefore likely to be postsynthetic. In latently infected cells, EBNA1 also regulates the expression of multiple EBV promoters located as many as 10 kbp away. EBNA1 supports replication and transcription through binding to oriP; both the ability of EBNA1 to bind to DNA and the integrity of its binding sites in oriP are required. However, DNA binding by EBNA1 is not sufficient to support replication or transcription, indicating that an additional activity (or activities) is required. EBNA1 links DNAs to which it binds and can form a loop between the two subelements of oriP, the family of repeats and the region of dyad symmetry, each of which contains multiple binding sites for EBNA1. We have constructed a set of derivatives of EBNA1 which contain both, one, or neither of its linking regions in various contexts. Analyses of these derivatives demonstrate that the linking regions of EBNA1 are essential for its support of replication and transcription and that the ability of derivatives of EBNA1 to link DNAs correlates strongly with their support of these activities in cells. These findings indicate that protein-protein associations of the linking regions of EBNA1 underlie its long-range contributions to replication and transcription.
Figures






References
-
- Aiyar A, Sugden B. Fusions between EBNA-1 of EBV and the large T-antigen of SV40 replicate their cognate origins. J Biol Chem. 1998;273:33073–33081. - PubMed
-
- Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E, Wahl G M, Epner E M. Participation of the human beta-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
