Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria
- PMID: 10207067
- PMCID: PMC84136
- DOI: 10.1128/MCB.19.5.3435
Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria
Abstract
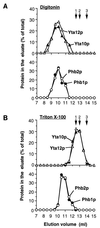
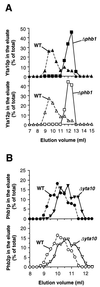
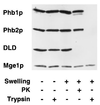
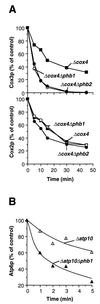
Prohibitins comprise a protein family in eukaryotic cells with potential roles in senescence and tumor suppression. Phb1p and Phb2p, members of the prohibitin family in Saccharomyces cerevisiae, have been implicated in the regulation of the replicative life span of the cells and in the maintenance of mitochondrial morphology. The functional activities of these proteins, however, have not been elucidated. We demonstrate here that prohibitins regulate the turnover of membrane proteins by the m-AAA protease, a conserved ATP-dependent protease in the inner membrane of mitochondria. The m-AAA protease is composed of the homologous subunits Yta10p (Afg3p) and Yta12p (Rca1p). Deletion of PHB1 or PHB2 impairs growth of Deltayta10 or Deltayta12 cells but does not affect cell growth in the presence of the m-AAA protease. A prohibitin complex with a native molecular mass of approximately 2 MDa containing Phb1p and Phb2p forms a supercomplex with the m-AAA protease. Proteolysis of nonassembled inner membrane proteins by the m-AAA protease is accelerated in mitochondria lacking Phb1p or Phb2p, indicating a negative regulatory effect of prohibitins on m-AAA protease activity. These results functionally link members of two conserved protein families in eukaryotes to the degradation of membrane proteins in mitochondria.
Figures


References
-
- Ackermann S H, Tzagoloff A. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1-F0 complex. J Biol Chem. 1990;265:9952–9959. - PubMed
-
- Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12-complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. - PubMed
-
- Brodsky J L, McCracken A A. ER-associated and proteasome-mediated protein degradation: how two topologically restricted events came together. Trends Cell Biol. 1997;7:151–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
