Stem-loop binding protein facilitates 3'-end formation by stabilizing U7 snRNP binding to histone pre-mRNA
- PMID: 10207079
- PMCID: PMC84148
- DOI: 10.1128/MCB.19.5.3561
Stem-loop binding protein facilitates 3'-end formation by stabilizing U7 snRNP binding to histone pre-mRNA
Abstract
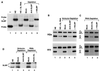
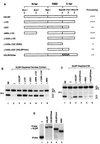
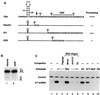
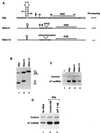
The 3' end of histone mRNA is formed by an endonucleolytic cleavage of the primary transcript after a conserved stem-loop sequence. The cleavage reaction requires at least two trans-acting factors: the stem-loop binding protein (SLBP), which binds the stem-loop sequence, and the U7 snRNP that interacts with a sequence downstream from the cleavage site. Removal of SLBP from a nuclear extract abolishes 3'-end processing, and the addition of recombinant SLBP restores processing activity of the depleted extract. To determine the regions of human SLBP necessary for 3' processing, various deletion mutants of the protein were tested for their ability to complement the SLBP-depleted extract. The entire N-terminal domain and the majority of the C-terminal domain of human SLBP are dispensable for processing. The minimal protein that efficiently supports cleavage of histone pre-mRNA consists of 93 amino acids containing the 73-amino-acid RNA-binding domain and 20 amino acids located immediately next to its C terminus. Replacement of these 20 residues with an unrelated sequence in the context of the full-length SLBP reduces processing >90%. Coimmunoprecipitation experiments with the anti-SLBP antibody demonstrated that SLBP and U7 snRNP form a stable complex only in the presence of pre-mRNA substrates containing a properly positioned U7 snRNP binding site. One role of SLBP is to stabilize the interaction of the histone pre-mRNA with U7 snRNP.
Figures


References
-
- Bond U M, Yario T A, Steitz J A. Multiple processing-defective mutations in a mammalian histone premessenger RNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes Dev. 1991;5:1709–1722. - PubMed
-
- Cotten M, Oberhauser B, Brunar H, Holzner A, Issakides G, Noe C R, Schaffner G, Wagner E, Birnstiel M L. 2′-O-Methyl, 2′-O-ethyl oligoribonucleotides and phosphorothioate oligodeoxyribonucleotides as inhibitors of the in vitro U7 snRNP-dependent mRNA processing event. Nucleic Acids Res. 1991;19:2629–2635. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
