Distinct mechanisms of activation of Stat1 and Stat3 by platelet-derived growth factor receptor in a cell-free system
- PMID: 10207096
- PMCID: PMC84192
- DOI: 10.1128/MCB.19.5.3727
Distinct mechanisms of activation of Stat1 and Stat3 by platelet-derived growth factor receptor in a cell-free system
Abstract
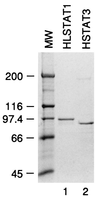
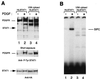
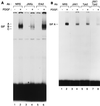
Ligand-dependent activation of the platelet-derived growth factor receptor (PDGFR) in fibroblasts in culture leads to the activation of the JAK family of protein-tyrosine kinases and of the transcription factors Stat1 and Stat3. To determine the biochemical mechanism of STAT activation by PDGFR, we devised a cell-free system composed of a membrane fraction from cells overexpressing PDGFR. When supplemented with crude cytosol, the membrane fraction supported PDGF- and ATP-dependent activation of both Stat1 and Stat3. However, the extent of Stat3 activation differed depending on the source of the cytosolic fraction. Using purified recombinant STAT proteins produced in Escherichia coli, we found that Stat1 could be activated by immunopurified PDGFR and showed no additional requirement for membrane- or cytosol-derived proteins. In contrast, activation of Stat3 exhibited a strong requirement for the cytosolic fraction. The activity present in the cytosolic fraction could be depleted with antibodies to JAK proteins. We conclude that the mechanisms of activation of Stat1 and Stat3 by PDGFR are distinct. Stat1 activation appears to result from a direct interaction with the receptor, whereas Stat3 activation additionally requires JAK proteins.
Figures







References
-
- Aman M J, Leonard W J. Cytokine signaling: cytokine-inducible signaling inhibitors. Curr Biol. 1997;7:784–788. - PubMed
-
- Bach E A, Auguet M, Schreiber R D. The IFNγ receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. - PubMed
-
- Briscoe J, Kohlhuber F, Müller M. JAKs and STATs branch out. Trends Cell Biol. 1996;6:336–340. - PubMed
-
- Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. - PubMed
-
- Darnell J J, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
