ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose
- PMID: 10207099
- PMCID: PMC84202
- DOI: 10.1128/MCB.19.5.3760
ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose
Abstract
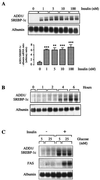
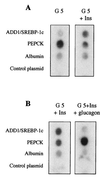
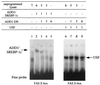
The transcription of genes encoding proteins involved in the hepatic synthesis of lipids from glucose is strongly stimulated by carbohydrate feeding. It is now well established that in the liver, glucose is the main activator of the expression of this group of genes, with insulin having only a permissive role. While ADD1/SREBP-1 has been implicated in lipogenic gene expression through temporal association with food intake and ectopic gain-of-function experiments, no genetic evidence for a requirement for this factor in glucose-mediated gene expression has been established. We show here that the transcription of ADD1/SREBP-1c in primary cultures of hepatocytes is controlled positively by insulin and negatively by glucagon and cyclic AMP, establishing a link between this transcription factor and carbohydrate availability. Using adenovirus-mediated transfection of a powerful dominant negative form of ADD1/SREBP-1c in rat hepatocytes, we demonstrate that this factor is absolutely necessary for the stimulation by glucose of L-pyruvate kinase, fatty acid synthase, S14, and acetyl coenzyme A carboxylase gene expression. These results demonstrate that ADD1/SREBP-1c plays a crucial role in mediating the expression of lipogenic genes induced by glucose and insulin.
Figures





References
-
- Brown M S, Goldstein J L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. - PubMed
-
- Chomczynski P, Sacchi N. Single step method of RNA isolation by acid guanidinium thiocyanate-phenol-choroform extraction. Anal Biochem. 1987;162:156–159. - PubMed
-
- Coupé C, Perdereau D, Ferré P, Hitier Y, Narkewicz M, Girard J. Lipogenic enzyme activities and mRNA in rat adipose tissue at weaning. Am J Physiol. 1990;258:E126–E133. - PubMed
-
- Doiron B, Cuif M H, Kahn A, Diaz-Guerra M J. Respective roles of glucose, fructose, and insulin in the regulation of the liver-specific pyruvate kinase gene promoter. J Biol Chem. 1994;269:10213–10216. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
