TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mutations
- PMID: 10207100
- PMCID: PMC84205
- DOI: 10.1128/MCB.19.5.3769
TAFII40 protein is encoded by the e(y)1 gene: biological consequences of mutations
Abstract
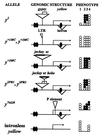
The enhancer of yellow 1 gene, e(y)1, of Drosophila melanogaster has been cloned and demonstrated to encode the TAFII40 protein. The e(y)1 gene is expressed in females much more strongly than in males due to the accumulation of e(y)1 mRNA in the ovaries. Two different e(y)1 mutations have been obtained. The e(y)1(ul) mutation, induced by the insertion of Stalker into the coding region, leads to the replacement of 25 carboxy-terminal amino acids by 17 amino acids encoded by the Stalker sequences and to a decrease of the e(y)1 transcription level. The latter is the main cause of dramatic underdevelopment of the ovaries and sterility of females bearing the e(y)1 mutation. This follows from the restoration of female fertility upon transformation of e(y)1(u1) flies with a construction synthesizing the mutant protein. The e(y)1(P1) mutation induced by P element insertion into the transcribed nontranslated region of the gene has almost no influence on the phenotype of flies. However, in combination with the phP1 mutation, which leads to a strong P element-mediated suppression of e(y)1 transcription, this mutation is lethal. Genetic studies of the e(y)1(u1) mutation revealed a sensitivity of the yellow and white expression to the TAFII40/e(y)1 level. The su(Hw)-binding region, Drosophila insulator, stabilizes the expression of the white gene and makes it independent of the e(y)1(u1) mutation.
Figures






References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Apone L M, Virbasius C-M A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. - PubMed
-
- Ashburner M. Drosophila: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989.
-
- Belenkaya T, Barseguyan K, Hovhannisayn H, Biryukova I, Kochieva E Z, Georgiev P. P element sequences can compensate a deletion of the yellow regulatory region in Drosophila melanogaster. Mol Gen Genet. 1998;259:79–87. - PubMed
-
- Belenkaya T, Soldatov A, Nabirochkina E, Birjukova I, Georgieva S, Georgiev P. The allele of the polyhomeotic gene induced by P element insertion encodes a new chimeric protein, that negatively regulates the expression of P-induced alleles in the yellow locus of Drosophila melanogaster. Genetics. 1998;150:687–697. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
