Molecular determinants of the estrogen receptor-coactivator interface
- PMID: 10207113
- PMCID: PMC84247
- DOI: 10.1128/MCB.19.5.3895
Molecular determinants of the estrogen receptor-coactivator interface
Abstract
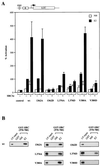
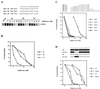
Transcriptional activation by the estrogen receptor is mediated through its interaction with coactivator proteins upon ligand binding. By systematic mutagenesis, we have identified a group of conserved hydrophobic residues in the ligand binding domain that are required for binding the p160 family of coactivators. Together with helix 12 and lysine 366 at the C-terminal end of helix 3, they form a hydrophobic groove that accommodates an LXXLL motif, which is essential for mediating coactivator binding to the receptor. Furthermore, we demonstrated that the high-affinity binding of motif 2, conserved in the p160 family, is due to the presence of three basic residues N terminal to the core LXXLL motif. The recruitment of p160 coactivators to the estrogen receptor is therefore likely to depend not only on the LXXLL motif making hydrophobic interactions with the docking surface on the receptor, but also on adjacent basic residues, which may be involved in the recognition of charged residues on the receptor to allow the initial docking of the motif.
Figures




Similar articles
-
Selection of estrogen receptor beta- and thyroid hormone receptor beta-specific coactivator-mimetic peptides using recombinant peptide libraries.Mol Endocrinol. 2000 May;14(5):605-22. doi: 10.1210/mend.14.5.0445. Mol Endocrinol. 2000. PMID: 10809226
-
Coactivator peptides have a differential stabilizing effect on the binding of estrogens and antiestrogens with the estrogen receptor.Mol Endocrinol. 1999 Nov;13(11):1912-23. doi: 10.1210/mend.13.11.0373. Mol Endocrinol. 1999. PMID: 10551784
-
Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation.Genes Dev. 1998 Nov 1;12(21):3357-68. doi: 10.1101/gad.12.21.3357. Genes Dev. 1998. PMID: 9808623 Free PMC article.
-
Structure-function evaluation of ER alpha and beta interplay with SRC family coactivators. ER selective ligands.Biochemistry. 2001 Jun 12;40(23):6756-65. doi: 10.1021/bi010379h. Biochemistry. 2001. PMID: 11389589
-
The coactivator LXXLL nuclear receptor recognition motif.J Pept Res. 2004 Mar;63(3):207-12. doi: 10.1111/j.1399-3011.2004.00126.x. J Pept Res. 2004. PMID: 15049832 Review.
Cited by
-
Aromatase-null mice expressing enhanced green fluorescent protein in germ cells provide a model system to assess estrogen-dependent ovulatory responses.Transgenic Res. 2014 Apr;23(2):293-302. doi: 10.1007/s11248-013-9771-y. Epub 2013 Nov 22. Transgenic Res. 2014. PMID: 24272335
-
LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1.Mol Cell Biol. 2003 Jan;23(1):238-49. doi: 10.1128/MCB.23.1.238-249.2003. Mol Cell Biol. 2003. PMID: 12482977 Free PMC article.
-
The role of androgen receptor mutations in prostate cancer progression.Curr Genomics. 2009 Mar;10(1):18-25. doi: 10.2174/138920209787581307. Curr Genomics. 2009. PMID: 19721807 Free PMC article.
-
The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes.Mol Cell Biol. 2000 Apr;20(8):2718-26. doi: 10.1128/MCB.20.8.2718-2726.2000. Mol Cell Biol. 2000. PMID: 10733574 Free PMC article.
-
T:G mismatch-specific thymine-DNA glycosylase (TDG) as a coregulator of transcription interacts with SRC1 family members through a novel tyrosine repeat motif.Nucleic Acids Res. 2005 Nov 10;33(19):6393-404. doi: 10.1093/nar/gki940. Print 2005. Nucleic Acids Res. 2005. PMID: 16282588 Free PMC article.
References
-
- Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
-
- Beato M, Herrlich P, Schutz G. Steroid-hormone receptors—many actors in search of a plot. Cell. 1995;83:851–857. - PubMed
-
- Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal-structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. - PubMed
-
- Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
