Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis
- PMID: 10209038
- PMCID: PMC2193031
- DOI: 10.1084/jem.189.8.1207
Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis
Abstract
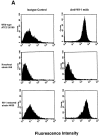
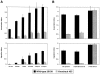
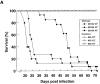
Systemic fungal infections are becoming more common and difficult to treat, yet the pathogenesis of these infectious diseases remains poorly understood. In many cases, pathogenicity can be attributed to the ability of the fungi to adhere to target tissues, but the lack of tractable genetic systems has limited progress in understanding and interfering with the offending fungal products. In Blastomyces dermatitidis, the agent of blastomycosis, a respiratory and disseminated mycosis of people and animals worldwide, expression of the putative adhesin encoded by the WI-1 gene was investigated as a possible virulence factor. DNA-mediated gene transfer was used to disrupt the WI-1 locus by allelic replacement, resulting in impaired binding and entry of yeasts into macrophages, loss of adherence to lung tissue, and abolishment of virulence in mice; each of these properties was fully restored after reconstitution of WI-1 by means of gene transfer. These findings establish the pivotal role of WI-1 in adherence and virulence of B. dermatitidis yeasts. To our knowledge, they offer the first example of a genetically proven virulence determinant among systemic dimorphic fungi, and underscore the value of reverse genetics for studies of pathogenesis in these organisms.
Figures






References
-
- Sternberg S. The emerging fungal threat. Science. 1994;266:1632–1634. - PubMed
-
- Ajello, L. 1970. Comparative study of pulmonary mycoses in the United States and Canada. In Proceedings of the Pan American Health Organization International Symposium on the Mycoses. Pan American Sanitary Bureau Scientific Publication No. 205, Washington, D.C. 3–10.
-
- Struhl K. The new yeast genetics. Nature. 1983;305:391–397. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

