Extended duration of DH-JH rearrangement in immunoglobulin heavy chain transgenic mice: implications for regulation of allelic exclusion
- PMID: 10209046
- PMCID: PMC2193035
- DOI: 10.1084/jem.189.8.1295
Extended duration of DH-JH rearrangement in immunoglobulin heavy chain transgenic mice: implications for regulation of allelic exclusion
Abstract
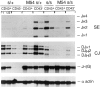
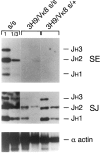
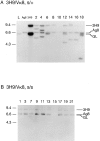
Here we show that suppression of VH-DJH rearrangement in mice bearing a mu heavy (H) chain transgene (mu-tg mice) is associated with an extended period of DH-JH rearrangement, the first step of Immunoglobulin H chain gene rearrangement. Whereas DH-JH rearrangement is normally initiated and completed at the pro-B cell stage, in mu-tg mice it continues beyond this stage and occurs most frequently at the small (late) pre-B stage. Despite ongoing DH-JH rearrangement in late pre-B cells of mu-tg mice, VH-DJH rearrangement is not detectable in these cells. We infer that the lack of VH-DJH rearrangement primarily reflects tg-induced acceleration of B cell differentiation past the stage at which rearrangement of VH elements is permissible. In support of this inference, we find that the normal representation of early B lineage subsets is markedly altered in mu-tg mice. We suggest that the effect of a productive VH-DJH rearrangement at an endogenous H chain allele may be similar to that of a mu-tg; i.e., cells that make a productive VH-DJH rearrangement on the first attempt rapidly progress to a developmental stage that precludes VH-DJH rearrangement at the other allele (allelic exclusion).
Figures



Similar articles
-
Biased reading frames of pre-existing DH--JH coding joints and preferential nucleotide insertions at VH--DJH signal joints of excision products of immunoglobulin heavy chain gene rearrangements.EMBO J. 1992 Dec;11(13):4869-75. doi: 10.1002/j.1460-2075.1992.tb05593.x. EMBO J. 1992. PMID: 1464314 Free PMC article.
-
The DJH complex remains active in recombination to VH segments after the loss of mu-chain expression in mu-positive pre-B cells.J Immunol. 1989 May 15;142(10):3652-6. J Immunol. 1989. PMID: 2497179
-
Ordered rearrangement of immunoglobulin heavy chain variable region segments.EMBO J. 1984 Jun;3(6):1209-19. doi: 10.1002/j.1460-2075.1984.tb01955.x. EMBO J. 1984. PMID: 6086308 Free PMC article.
-
Control of the sizes and contents of precursor B cell repertoires in bone marrow.Ciba Found Symp. 1997;204:172-82; discussion 182-6. doi: 10.1002/9780470515280.ch12. Ciba Found Symp. 1997. PMID: 9107420 Review.
-
Immunoglobulin VH gene analysis in rat: most marginal zone B cells express germline encoded VH genes and are ligand selected.Curr Top Microbiol Immunol. 2000;252:107-17. doi: 10.1007/978-3-642-57284-5_12. Curr Top Microbiol Immunol. 2000. PMID: 11125468 Review. No abstract available.
Cited by
-
Biphenotypic plasma cell myeloma: two cases of plasma cell neoplasm with a coexpression of kappa and lambda light chains.Int J Clin Exp Pathol. 2015 Jul 1;8(7):8536-44. eCollection 2015. Int J Clin Exp Pathol. 2015. PMID: 26339430 Free PMC article.
-
Complete cis Exclusion upon Duplication of the Eμ Enhancer at the Immunoglobulin Heavy Chain Locus.Mol Cell Biol. 2015 Jul;35(13):2231-41. doi: 10.1128/MCB.00294-15. Mol Cell Biol. 2015. PMID: 25896912 Free PMC article.
-
Multiple, conserved cryptic recombination signals in VH gene segments: detection of cleavage products only in pro B cells.J Exp Med. 2007 Dec 24;204(13):3195-208. doi: 10.1084/jem.20071224. Epub 2007 Dec 3. J Exp Med. 2007. PMID: 18056287 Free PMC article.
-
Differential effect of an Ig mu transgene on development of pre-B cells in fetal and adult SCID mice.Proc Natl Acad Sci U S A. 1999 Oct 12;96(21):11952-7. doi: 10.1073/pnas.96.21.11952. Proc Natl Acad Sci U S A. 1999. PMID: 10518557 Free PMC article.
-
Chromosomal translocation t(11;14) and p53 deletion induced by the CRISPR/Cas9 system in normal B cell-derived iPS cells.Sci Rep. 2021 Mar 4;11(1):5216. doi: 10.1038/s41598-021-84628-5. Sci Rep. 2021. PMID: 33664418 Free PMC article.
References
-
- Coleclough CR, Perry RP, Karjalainen K, Weigert M. Aberrant rearrangements contribute significantly to the allelic exclusion of immunoglobulin gene expression. Nature. 1981;290:372–377. - PubMed
-
- Alt FW, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MuLV–transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

