ebi regulates epidermal growth factor receptor signaling pathways in Drosophila
- PMID: 10215623
- PMCID: PMC316643
- DOI: 10.1101/gad.13.8.954
ebi regulates epidermal growth factor receptor signaling pathways in Drosophila
Abstract
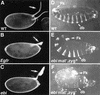
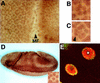
ebi regulates the epidermal growth factor receptor (EGFR) signaling pathway at multiple steps in Drosophila development. Mutations in ebi and Egfr lead to similar phenotypes and show genetic interactions. However, ebi does not show genetic interactions with other RTKs (e.g., torso) or with components of the canonical Ras/MAP kinase pathway. ebi encodes an evolutionarily conserved protein with a unique amino terminus, distantly related to F-box sequences, and six tandemly arranged carboxy-terminal WD40 repeats. The existence of closely related proteins in yeast, plants, and humans suggests that ebi functions in a highly conserved biochemical pathway. Proteins with related structures regulate protein degradation. Similarly, in the developing eye, ebi promotes EGFR-dependent down-regulation of Tramtrack88, an antagonist of neuronal development.
Figures








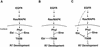
Similar articles
-
An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling.Cell. 2002 Sep 6;110(5):625-37. doi: 10.1016/s0092-8674(02)00875-9. Cell. 2002. PMID: 12230979
-
A role for Ebi in neuronal cell cycle control.EMBO J. 2000 Oct 16;19(20):5376-86. doi: 10.1093/emboj/19.20.5376. EMBO J. 2000. PMID: 11032805 Free PMC article.
-
Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins.Genes Dev. 1996 Sep 15;10(18):2302-13. doi: 10.1101/gad.10.18.2302. Genes Dev. 1996. PMID: 8824589
-
New Insights from Drosophila into the Regulation of EGFR Signaling.Methods Mol Biol. 2017;1652:37-42. doi: 10.1007/978-1-4939-7219-7_2. Methods Mol Biol. 2017. PMID: 28791632 Review.
-
Complexity of EGF receptor signalling revealed in Drosophila.Curr Opin Genet Dev. 1998 Aug;8(4):407-11. doi: 10.1016/s0959-437x(98)80110-x. Curr Opin Genet Dev. 1998. PMID: 9729715 Review.
Cited by
-
A Drosophila model of multiple endocrine neoplasia type 2.Genetics. 2005 Nov;171(3):1057-81. doi: 10.1534/genetics.104.038018. Epub 2005 Jun 18. Genetics. 2005. PMID: 15965261 Free PMC article.
-
Phyllopod acts as an adaptor protein to link the sina ubiquitin ligase to the substrate protein tramtrack.Mol Cell Biol. 2002 Oct;22(19):6854-65. doi: 10.1128/MCB.22.19.6854-6865.2002. Mol Cell Biol. 2002. PMID: 12215542 Free PMC article.
-
Ebi, a Drosophila homologue of TBL1, regulates the balance between cellular defense responses and neuronal survival.Am J Neurodegener Dis. 2016 Mar 1;5(1):62-8. eCollection 2016. Am J Neurodegener Dis. 2016. PMID: 27073743 Free PMC article.
-
TBL1XR1 in physiological and pathological states.Am J Clin Exp Urol. 2015 Apr 25;3(1):13-23. eCollection 2015. Am J Clin Exp Urol. 2015. PMID: 26069883 Free PMC article. Review.
-
Lissencephaly 1 linking to multiple diseases: mental retardation, neurodegeneration, schizophrenia, male sterility, and more.Neuromolecular Med. 2006;8(4):547-65. doi: 10.1385/NMM:8:4:547. Neuromolecular Med. 2006. PMID: 17028375 Review.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bai C, Sen P, Hofman K, Ma L, Goebl M, Harper JW, Elledge S. Skp1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. - PubMed
-
- Banerjee U, Renfranz PJ, Pollock JA, Benzer S. Molecular characterization and expression of sevenless, a gene involved in neuronal pattern formation in the Drosophila eye. Cell. 1987;49:281–291. - PubMed
-
- Basler K, Christen B, Hafen E. Ligand-independent activation of the sevenless Receptor tyrosine kinase changes the fate of cells in the developing Drosophila eye. Cell. 1991;64:1069–1081. - PubMed
-
- Bronner G, Chu-LaGraff Q, Doe CQ, Cohen B, Weigel D, Taubert H, Jäckle H. Sp1/egr-like zinc-finger protein required for endoderm specification and germ-layer formation in Drosophila. Nature. 1994;369:664–668. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
