The global nitrogen regulator NtcA regulates transcription of the signal transducer PII (GlnB) and influences its phosphorylation level in response to nitrogen and carbon supplies in the Cyanobacterium synechococcus sp. strain PCC 7942
- PMID: 10217756
- PMCID: PMC93707
- DOI: 10.1128/JB.181.9.2697-2702.1999
The global nitrogen regulator NtcA regulates transcription of the signal transducer PII (GlnB) and influences its phosphorylation level in response to nitrogen and carbon supplies in the Cyanobacterium synechococcus sp. strain PCC 7942
Abstract
The PII protein is encoded by a unique glnB gene in Synechococcus sp. strain PCC 7942. Its expression has been analyzed in the wild type and in NtcA-null mutant cells grown under different conditions of nitrogen and carbon supply. RNA-DNA hybridization experiments revealed the presence of one transcript species 680 nucleotides long, whatever the nutrient conditions tested. A second transcript species, 620 nucleotides long, absent in the NtcA null mutant, was observed in wild-type cells that were nitrogen starved for 2 h under both high and low CO2 and in the presence of nitrate under a high CO2 concentration. Primer extension analysis indicated that the two transcript species are generated from two tandem promoters, a sigma70 Escherichia coli-type promoter and an NtcA-dependent promoter, located 120 and 53 nucleotides, respectively, from the glnB initiation codon. The NtcA-dependent promoter is up-regulated under the conditions mentioned above, while the sigma70 E. coli-type promoter displays constitutive levels of transcripts in the NtcA null mutant and slightly different levels in the wild-type cells, depending on the nitrogen and carbon supplies. In general, a good correlation between the amounts of the two transcript species and that of the PII protein was observed, as revealed by immunodetection with specific antibodies. The phosphorylation level of PII in the wild type is inversely correlated with nitrogen availability and directly correlated with higher CO2 concentration. This regulation is correspondingly less stringent in the NtcA null mutant cells. In contrast, the dephosphorylation of PII is NtcA independent.
Figures





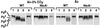

References
-
- Amar M, Patriarca E J, Manco G, Bernard P, Riccio A, Lamberti A, Defez R, Iaccarino M. Regulation of nitrogen metabolism is altered in a glnB mutant strain of Rhizobium leguminosarum. Mol Microbiol. 1994;11:685–693. - PubMed
-
- Chiurazzi M, Iaccarino M. Transcriptional analysis of the glnB-glnA region of Rhizobium leguminosarum biovar viciae. Mol Microbiol. 1990;4:1727–1735. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

