Pullulanase type I from Fervidobacterium pennavorans Ven5: cloning, sequencing, and expression of the gene and biochemical characterization of the recombinant enzyme
- PMID: 10224005
- PMCID: PMC91302
- DOI: 10.1128/AEM.65.5.2084-2091.1999
Pullulanase type I from Fervidobacterium pennavorans Ven5: cloning, sequencing, and expression of the gene and biochemical characterization of the recombinant enzyme
Abstract
The gene encoding the type I pullulanase from the extremely thermophilic anaerobic bacterium Fervidobacterium pennavorans Ven5 was cloned and sequenced in Escherichia coli. The pulA gene from F. pennavorans Ven5 had 50.1% pairwise amino acid identity with pulA from the anaerobic hyperthermophile Thermotoga maritima and contained the four regions conserved among all amylolytic enzymes. The pullulanase gene (pulA) encodes a protein of 849 amino acids with a 28-residue signal peptide. The pulA gene was subcloned without its signal sequence and overexpressed in E. coli under the control of the trc promoter. This clone, E. coli FD748, produced two proteins (93 and 83 kDa) with pullulanase activity. A second start site, identified 118 amino acids downstream from the ATG start site, with a Shine-Dalgarno-like sequence (GGAGG) and TTG translation initiation codon was mutated to produce only the 93-kDa protein. The recombinant purified pullulanases (rPulAs) were optimally active at pH 6 and 80 degrees C and had a half-life of 2 h at 80 degrees C. The rPulAs hydrolyzed alpha-1,6 glycosidic linkages of pullulan, starch, amylopectin, glycogen, alpha-beta-limited dextrin. Interestingly, amylose, which contains only alpha-1,4 glycosidic linkages, was not hydrolyzed by rPulAs. According to these results, the enzyme is classified as a debranching enzyme, pullulanase type I. The extraordinary high substrate specificity of rPulA together with its thermal stability makes this enzyme a good candidate for biotechnological applications in the starch-processing industry.
Figures

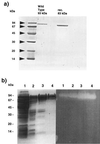
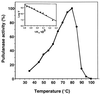

References
-
- Albertson G D, McHale R H, Gibbs M D, Bergquist P L. Cloning and sequence of a type I pullulanase from an extremely thermophilic anaerobic bacterium, Caldicellulosiruptor saccharolyticus. Biochim Biophys Acta. 1997;1354:35–39. - PubMed
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Antranikian, G., P. L. Jørgensen, M. Wümpelmann, and S. T. Jørgensen. 20 February 1992. Novel thermostable pullulanases. Patent WO 92/02614.
-
- Bernfeld P. Amylases α and β. Methods Enzymol. 1955;1:149–155.
-
- Bibel M, Brettl C, Gosslar U, Kriegshäuser G, Liebl W. Isolation and analysis of genes for amylolytic enzymes of the hyperthermophilic bacterium Thermotoga maritima. FEMS Microbiol Lett. 1998;158:9–15. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

