Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus
- PMID: 10224020
- PMCID: PMC91317
- DOI: 10.1128/AEM.65.5.2202-2208.1999
Analysis of 16S-23S rRNA intergenic spacer regions of Vibrio cholerae and Vibrio mimicus
Abstract
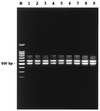
Vibrio cholerae identification based on molecular sequence data has been hampered by a lack of sequence variation from the closely related Vibrio mimicus. The two species share many genes coding for proteins, such as ctxAB, and show almost identical 16S DNA coding for rRNA (rDNA) sequences. Primers targeting conserved sequences flanking the 3' end of the 16S and the 5' end of the 23S rDNAs were used to amplify the 16S-23S rRNA intergenic spacer regions of V. cholerae and V. mimicus. Two major (ca. 580 and 500 bp) and one minor (ca. 750 bp) amplicons were consistently generated for both species, and their sequences were determined. The largest fragment contains three tRNA genes (tDNAs) coding for tRNAGlu, tRNALys, and tRNAVal, which has not previously been found in bacteria examined to date. The 580-bp amplicon contained tDNAIle and tDNAAla, whereas the 500-bp fragment had single tDNA coding either tRNAGlu or tRNAAla. Little variation, i.e., 0 to 0.4%, was found among V. cholerae O1 classical, O1 El Tor, and O139 epidemic strains. Slightly more variation was found against the non-O1/non-O139 serotypes (ca. 1% difference) and V. mimicus (2 to 3% difference). A pair of oligonucleotide primers were designed, based on the region differentiating all of V. cholerae strains from V. mimicus. The PCR system developed was subsequently evaluated by using representatives of V. cholerae from environmental and clinical sources, and of other taxa, including V. mimicus. This study provides the first molecular tool for identifying the species V. cholerae.
Figures



References
-
- Albert M J, Siddique A K, Islam M S, Faruque A S, Ansaruzzaman M, Faruque S M, Sack R B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993;341:704. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

