The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity
- PMID: 10224280
- PMCID: PMC2193055
- DOI: 10.1084/jem.189.9.1399
The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity
Abstract
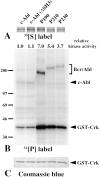
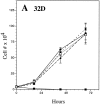
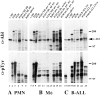
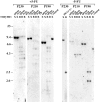
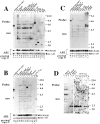
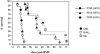
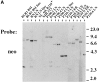
The product of the Philadelphia chromosome (Ph) translocation, the BCR/ABL oncogene, exists in three principal forms (P190, P210, and P230 BCR/ABL) that are found in distinct forms of Ph-positive leukemia, suggesting the three proteins have different leukemogenic activity. We have directly compared the tyrosine kinase activity, in vitro transformation properties, and in vivo leukemogenic activity of the P190, P210, and P230 forms of BCR/ABL. P230 exhibited lower intrinsic tyrosine kinase activity than P210 and P190. Although all three oncogenes transformed both myeloid (32D cl3) and lymphoid (Ba/F3) interleukin (IL)-3-dependent cell lines to become independent of IL-3 for survival and growth, their ability to stimulate proliferation of Ba/F3 lymphoid cells differed and correlated directly with tyrosine kinase activity. In a murine bone marrow transduction/transplantation model, the three forms of BCR/ABL were equally potent in the induction of a chronic myeloid leukemia (CML)-like myeloproliferative syndrome in recipient mice when 5-fluorouracil (5-FU)-treated donors were used. Analysis of proviral integration showed the CML-like disease to be polyclonal and to involve multiple myeloid and B lymphoid lineages, implicating a primitive multipotential target cell. Secondary transplantation revealed that only certain minor clones gave rise to day 12 spleen colonies and induced disease in secondary recipients, suggesting heterogeneity among the target cell population. In contrast, when marrow from non- 5-FU-treated donors was used, a mixture of CML-like disease, B lymphoid acute leukemia, and macrophage tumors was observed in recipients. P190 BCR/ABL induced lymphoid leukemia with shorter latency than P210 or P230. The lymphoid leukemias and macrophage tumors had provirus integration patterns that were oligo- or monoclonal and limited to the tumor cells, suggesting a lineage-restricted target cell with a requirement for additional events in addition to BCR/ABL transduction for full malignant transformation. These results do not support the hypothesis that P230 BCR/ABL induces a distinct and less aggressive form of CML in humans, and suggest that the rarity of P190 BCR/ABL in human CML may reflect infrequent BCR intron 1 breakpoints during the genesis of the Ph chromosome in stem cells, rather than intrinsic differences in myeloid leukemogenicity between P190 and P210.
Figures






References
-
- Fainstein E, Marcelle C, Rosner A, Canaani E, Gale RP, Dreazen O, Smith SD, Croce CM. A new fused transcript in Philadelphia chromosome positive acute lymphocytic leukaemia. Nature. 1987;330:386–388. - PubMed
-
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. - PubMed
-
- Saglio G, Guerrasio A, Rosso C, Zaccaria A, Tassinari A, Serra A, Rege-Cambrin G, Mazza U, Gavosto F. New type of Bcr/Abl junction in Philadelphia chromosome-positive chronic myelogenous leukemia. Blood. 1990;76:1819–1824. - PubMed
-
- Van Etten, R.A. 1993. The molecular pathogenesis of the Philadelphia-positive leukemias: implications for diagnosis and therapy. In Leukemia: Advances in Treatment and Research. E.J. Freireich and H. Kantarjian, editors. Kluwer Academic Publishers, Norwood, MA. 294–325. - PubMed
-
- Melo JV. The diversity of BCR-ABL fusion proteins and their relationship to leukemia phenotype. Blood. 1996;88:2375–2384. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

