Functional expression of Nramp1 in vitro in the murine macrophage line RAW264.7
- PMID: 10225878
- PMCID: PMC115961
- DOI: 10.1128/IAI.67.5.2225-2232.1999
Functional expression of Nramp1 in vitro in the murine macrophage line RAW264.7
Abstract
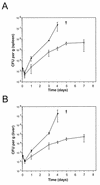
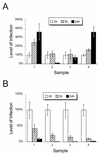
Mutations at the Nramp1 locus in vivo cause susceptibility to infection by unrelated intracellular microbes. Nramp1 encodes an integral membrane protein abundantly expressed in the endosomal-lysosomal compartment of macrophages and is recruited to the phagosomal membrane following phagocytosis. The mechanism by which Nramp1 affects the biochemical properties of the phagosome to control microbial replication is unknown. To devise an in vitro assay for Nramp1 function, we introduced a wild-type Nramp1(G169) cDNA into RAW 264.7 macrophages (which bear a homozygous mutant Nramp1(D169) allele and thus are permissive to replication of specific intracellular parasites). Recombinant Nramp1 was expressed in a membranous compartment in RAW264.7 cells and was recruited to the membrane of Salmonella typhimurium and Yersinia enterocolitica containing phagosomes. Evaluation of the antibacterial activity of RAW264.7 transfectants showed that expression of the recombinant Nramp1 protein abrogated intracellular replication of S. typhimurium. Studies with a replication-defective S. typhimurium mutant suggest that this occurs through an enhanced bacteriostatic activity. The effect of Nramp1 expression was specific, since (i) it was not seen in RAW264.7 transfectants overexpressing the closely related Nramp2 protein, and (ii) control RAW264.7 cells, Nramp1, and Nramp2 transfectants could all efficiently kill a temperature-sensitive, replication-defective mutant of S. typhimurium. Finally, increased antibacterial activity of the Nramp1 RAW264.7 transfectants was linked to increased phagosomal acidification, a distinguishing feature of primary macrophages expressing a wild-type Nramp1 allele. Together, these results indicate that transfection of Nramp1 cDNAs in the RAW264.7 macrophage cell line can be used as a direct assay to study both Nramp1 function and mechanism of action as well as to identify structure-function relationships in this protein.
Figures


Similar articles
-
Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1 D169) mouse strains.J Immunol. 1996 Oct 15;157(8):3559-68. J Immunol. 1996. PMID: 8871656
-
Natural resistance to infection with intracellular pathogens: the Nramp1 protein is recruited to the membrane of the phagosome.J Exp Med. 1997 Feb 17;185(4):717-30. doi: 10.1084/jem.185.4.717. J Exp Med. 1997. PMID: 9034150 Free PMC article.
-
Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane.J Exp Med. 2000 Nov 6;192(9):1237-48. doi: 10.1084/jem.192.9.1237. J Exp Med. 2000. PMID: 11067873 Free PMC article.
-
Nramp1: a link between intracellular iron transport and innate resistance to intracellular pathogens.J Leukoc Biol. 1999 Nov;66(5):757-62. doi: 10.1002/jlb.66.5.757. J Leukoc Biol. 1999. PMID: 10577506 Review.
-
The Nramp1 protein and its role in resistance to infection and macrophage function.Proc Assoc Am Physicians. 1999 Jul-Aug;111(4):283-9. doi: 10.1046/j.1525-1381.1999.99236.x. Proc Assoc Am Physicians. 1999. PMID: 10417735 Review.
Cited by
-
Molecular Mechanism of Nramp-Family Transition Metal Transport.J Mol Biol. 2021 Aug 6;433(16):166991. doi: 10.1016/j.jmb.2021.166991. Epub 2021 Apr 16. J Mol Biol. 2021. PMID: 33865868 Free PMC article. Review.
-
Ribonucleotide reductases of Salmonella typhimurium: transcriptional regulation and differential role in pathogenesis.PLoS One. 2010 Jun 25;5(6):e11328. doi: 10.1371/journal.pone.0011328. PLoS One. 2010. PMID: 20593029 Free PMC article.
-
Salmonella enterica serovar typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence In vivo.Infect Immun. 2000 Aug;68(8):4485-91. doi: 10.1128/IAI.68.8.4485-4491.2000. Infect Immun. 2000. PMID: 10899846 Free PMC article.
-
Salmonella enterica infection stimulates macrophages to hemophagocytose.mBio. 2014 Dec 9;5(6):e02211. doi: 10.1128/mBio.02211-14. mBio. 2014. PMID: 25491357 Free PMC article.
-
Nramp1 expression by dendritic cells modulates inflammatory responses during Salmonella Typhimurium infection.Cell Microbiol. 2008 Aug;10(8):1646-61. doi: 10.1111/j.1462-5822.2008.01155.x. Epub 2008 Apr 7. Cell Microbiol. 2008. PMID: 18397382 Free PMC article.
References
-
- Abel L, Sanchez F O, Oberti J, Thuc N V, Hoa L V, Lap V D, Skamene E, Lagrange P H, Schurr E. Susceptibility to leprosy is linked to the human NRAMP1 gene. J Infect Dis. 1998;177:133–145. - PubMed
-
- Bellamy R, Ruwende C, Corrah T, McAdam K P, Whittle H C, Hill A V. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. - PubMed
-
- Blackwell J M, Roach T I A, Atkinson S E, Ajioka J W, Barton C H, Shaw M A. Genetic regulation of macrophage priming activation: the Lsh gene story. Immunol Lett. 1991;30:241–248. - PubMed
-
- Buschman E, Taniyama T, Nakamura R, Skamene E. Functional expression of the Bcg gene in macrophage. Res Immunol. 1989;140:793–797. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

