The Drosophila dCREB2 gene affects the circadian clock
- PMID: 10230797
- PMCID: PMC11772075
- DOI: 10.1016/s0896-6273(00)80736-9
The Drosophila dCREB2 gene affects the circadian clock
Abstract
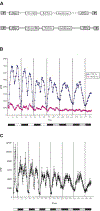
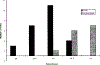
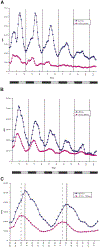
We report the role of dCREB2, the Drosophila homolog of CREB/CREM, in circadian rhythms. dCREB2 activity cycles with a 24 hr rhythm in flies, both in a light:dark cycle and in constant darkness. A mutation in dCREB2 shortens circadian locomotor rhythm in flies and dampens the oscillation of period, a known clock gene. Cycling dCREB2 activity is abolished in a period mutant, indicating that dCREB2 and Period affect each other and suggesting that the two genes participate in the same regulatory feedback loop. We propose that dCREB2 supports cycling of the Period/Timeless oscillator. These findings support CREB's role in mediating adaptive behavioral responses to a variey of environmental stimuli (stress, growth factors, drug addiction, circadian rhythms, and memory formation) in mammals and long-term memory formation and circadian rhythms in Drosophila.
Figures




Similar articles
-
Transgenic cAMP response element reporter flies for monitoring circadian rhythms.Methods Enzymol. 2005;393:302-15. doi: 10.1016/S0076-6879(05)93013-9. Methods Enzymol. 2005. PMID: 15817296
-
In vivo circadian oscillation of dCREB2 and NF-κB activity in the Drosophila nervous system.PLoS One. 2012;7(10):e45130. doi: 10.1371/journal.pone.0045130. Epub 2012 Oct 15. PLoS One. 2012. PMID: 23077489 Free PMC article.
-
Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks.J Biol Rhythms. 2001 Jun;16(3):205-15. doi: 10.1177/074873040101600303. J Biol Rhythms. 2001. PMID: 11407780
-
Genetics and molecular biology of rhythms in Drosophila and other insects.Adv Genet. 2003;48:1-280. doi: 10.1016/s0065-2660(03)48000-0. Adv Genet. 2003. PMID: 12593455 Review.
-
Circadian Rhythms and Sleep in Drosophila melanogaster.Genetics. 2017 Apr;205(4):1373-1397. doi: 10.1534/genetics.115.185157. Genetics. 2017. PMID: 28360128 Free PMC article. Review.
Cited by
-
Comparative approaches to the study of physiology: Drosophila as a physiological tool.Am J Physiol Regul Integr Comp Physiol. 2013 Feb;304(3):R177-88. doi: 10.1152/ajpregu.00084.2012. Epub 2012 Dec 5. Am J Physiol Regul Integr Comp Physiol. 2013. PMID: 23220476 Free PMC article. Review.
-
A genetic screen identifies putative targets and binding partners of CREB-binding protein in the developing Drosophila eye.Genetics. 2005 Dec;171(4):1655-72. doi: 10.1534/genetics.105.045450. Epub 2005 Jul 5. Genetics. 2005. PMID: 15998717 Free PMC article.
-
Overexpression of neprilysin reduces alzheimer amyloid-beta42 (Abeta42)-induced neuron loss and intraneuronal Abeta42 deposits but causes a reduction in cAMP-responsive element-binding protein-mediated transcription, age-dependent axon pathology, and premature death in Drosophila.J Biol Chem. 2008 Jul 4;283(27):19066-76. doi: 10.1074/jbc.M710509200. Epub 2008 May 7. J Biol Chem. 2008. PMID: 18463098 Free PMC article.
-
Central and peripheral clocks are coupled by a neuropeptide pathway in Drosophila.Nat Commun. 2017 May 30;8:15563. doi: 10.1038/ncomms15563. Nat Commun. 2017. PMID: 28555616 Free PMC article.
-
CREB regulation of BK channel gene expression underlies rapid drug tolerance.Genes Brain Behav. 2009 Jun;8(4):369-76. doi: 10.1111/j.1601-183X.2009.00479.x. Epub 2009 Feb 9. Genes Brain Behav. 2009. PMID: 19243452 Free PMC article.
References
-
- Allada R, White NE, So WV, Hall JC, and Rosbash M (1998). A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93, 791–804. - PubMed
-
- Amir S, and Stewart J (1996). Resetting of the circadian clock by a conditioned stimulus. Nature 379, 542–545. - PubMed
-
- Ausubel FM, Kingston RE, Moore DD, Seidman JG, Smith JA, and Struhl K, eds. (1989). Current Protocols in Molecular Biology, Sections 10.2 and 10.8 (New York: Wiley Interscience; ).
-
- Balsalobre A, Damiola F, and Schibler U (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937. - PubMed
-
- Borsook D, Konradi C, Falkowski O, Comb M, and Hyman SE (1994). Molecular mechanisms of stress-induced proenkephalin gene regulation: CREB interacts with the proenkephalin gene in the mouse hypothalamus and is phosphorylated in response to hyperosmolar stress. Mol. Endocrinol. 8, 240–248. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

