Genetics of graft-versus-host disease, I. A locus on chromosome 1 influences development of acute graft-versus-host disease in a major histocompatibility complex mismatched murine model
- PMID: 10233703
- PMCID: PMC2326746
- DOI: 10.1046/j.1365-2567.1999.00626.x
Genetics of graft-versus-host disease, I. A locus on chromosome 1 influences development of acute graft-versus-host disease in a major histocompatibility complex mismatched murine model
Abstract
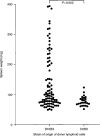
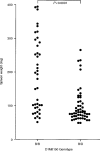
Graft-versus-host disease (GVHD) is the major complication occurring after bone marrow transplantation. The severity of GVHD varies widely, with this variation generally being attributed to variation in the degree of disparity between host and donor for minor histocompatibility antigens. However, it is also possible that other forms of polymorphism, such as polymorphisms in immune effector molecules, might play a significant role in determining GVHD severity. In order to investigate this hypothesis, we are studying the genetic factors that influence GVHD development in a murine model. We here report the first results of this analysis, which demonstrate that a locus on Chromosome 1 of the mouse, and possibly also a locus on Chromosome 4, exert considerable influence over the development of one aspect of acute GVHD - splenomegaly - in a parent-->F1 murine model. These results demonstrate that non-MHC genes can exert quite significant effects on the development of GVHD-associated pathology and that gene mapping can be used as a tool to identify these loci. Further analysis of such loci will allow identification of the mechanism whereby they influence GVHD and may lead in the future to improved selection of donors for human bone marrow transplantation.
Figures


References
-
- Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogen. 1997;24:1. - PubMed
-
- Bailly S, di Giovine FS, Blakemore AI, Duff GW. Genetic polymorphism of human interleukin-1 alpha. Eur J Immunol. 1993;23:1240. - PubMed
-
- Matesanz F, Alcina A. Glutamine and tetrapeptide repeat variations affect the biological activity of different mouse interleukin-2 alleles. Eur J Immunol. 1996;26:1675. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

