Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of early lytic cycle genes and synergy with the Epstein-Barr virus R transactivator
- PMID: 10233912
- PMCID: PMC112494
- DOI: 10.1128/JVI.73.6.4543-4551.1999
Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of early lytic cycle genes and synergy with the Epstein-Barr virus R transactivator
Abstract
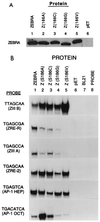
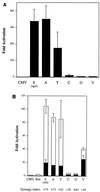
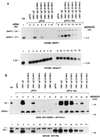
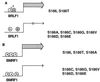
The ZEBRA protein mediates the switch between the latent and lytic life cycles of Epstein-Barr virus. Z(S186A), a point mutant in ZEBRA's basic domain in which serine 186 is changed to alanine, is unable to induce expression of lytic cycle mRNAs or proteins from the latent EBV genome even though it retains the ability to activate transcription from reporters bearing known ZEBRA-responsive promoters (A. L. Francis et al., J. Virol. 71:3054-3061, 1997). We now describe three distinct phenotypes of ZEBRA mutants bearing different amino acid substitutions at S186. These phenotypes are based on the capacity of the mutants to activate expression of the BRLF1 and BMRF1 genes, which are targets of ZEBRA's action, and to synergize with the BRLF1 gene product Rta (R transactivator) in activating expression of downstream genes. One mutant class, represented by Z(S186T), was similar to the wild type, although reduced in the capacity to activate BRLF1 and BMRF1 early lytic cycle genes from the latent virus. A second class, represented by Z(S186C) and Z(S186G), was impaired in transcriptional activation, unable to activate early lytic cycle products from the latent virus, and not rescued by overexpression of Rta. A third class, Z(S186A), although unable by itself to activate BRLF1 or other lytic cycle genes, synergized with Rta. Rta rescued the capacity of Z(S186A) to activate the BMRF1 early lytic cycle gene from the latent virus. All mutant classes bound to DNA in vitro, although their capacity to bind to different ZEBRA response elements varied. Serine 186 of ZEBRA is a critical residue that is required for the distinct activities of induction of BRLF1 expression and for synergy with Rta. Since only Z(S186T) among the mutants behaved similarly to the wild type, activation of BRLF1 likely requires phosphorylation of S186. However, since Z(S186A) could synergize with Rta, synergy with Rta does not appear to be dependent on phosphorylation of S186. S186 likely mediates DNA recognition on the BRLF1 promoter in the context of the latent virus, protein-protein interactions, or both. The Z(S186) mutants define the amino acid side chains required for these functions.
Figures


Similar articles
-
Role of the epstein-barr virus RTA protein in activation of distinct classes of viral lytic cycle genes.J Virol. 1999 Dec;73(12):9858-66. doi: 10.1128/JVI.73.12.9858-9866.1999. J Virol. 1999. PMID: 10559298 Free PMC article.
-
Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product.Virology. 1998 Nov 10;251(1):187-97. doi: 10.1006/viro.1998.9396. Virology. 1998. PMID: 9813214
-
Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta.J Virol. 2004 Jul;78(14):7634-44. doi: 10.1128/JVI.78.14.7634-7644.2004. J Virol. 2004. PMID: 15220438 Free PMC article.
-
Lytic cycle switches of oncogenic human gammaherpesviruses.Adv Cancer Res. 2007;97:81-109. doi: 10.1016/S0065-230X(06)97004-3. Adv Cancer Res. 2007. PMID: 17419942 Review.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
Cited by
-
Essential role of Rta in lytic DNA replication of Epstein-Barr virus.J Virol. 2013 Jan;87(1):208-23. doi: 10.1128/JVI.01995-12. Epub 2012 Oct 17. J Virol. 2013. PMID: 23077295 Free PMC article.
-
Mechanisms of T cell evasion by Epstein-Barr virus and implications for tumor survival.Front Immunol. 2023 Dec 21;14:1289313. doi: 10.3389/fimmu.2023.1289313. eCollection 2023. Front Immunol. 2023. PMID: 38179040 Free PMC article. Review.
-
Characterization of gammaherpesvirus 68 gene 50 transcription.J Virol. 2000 Feb;74(4):2029-37. doi: 10.1128/jvi.74.4.2029-2037.2000. J Virol. 2000. PMID: 10644377 Free PMC article.
-
Viral genome methylation differentially affects the ability of BZLF1 versus BRLF1 to activate Epstein-Barr virus lytic gene expression and viral replication.J Virol. 2013 Jan;87(2):935-50. doi: 10.1128/JVI.01790-12. Epub 2012 Nov 7. J Virol. 2013. PMID: 23135711 Free PMC article.
-
Methyl-dependent and spatial-specific DNA recognition by the orthologous transcription factors human AP-1 and Epstein-Barr virus Zta.Nucleic Acids Res. 2017 Mar 17;45(5):2503-2515. doi: 10.1093/nar/gkx057. Nucleic Acids Res. 2017. PMID: 28158710 Free PMC article.
References
-
- Adamson A L, Kenney S C. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology. 1998;251:187–197. - PubMed
-
- Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Stimulation of c-Jun activity by CBP: c-Jun residues Ser63/73 are required for CBP-induced stimulation in vivo and CBP binding in vitro. Oncogene. 1995;11:2509–2514. - PubMed
-
- Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3:488–499. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

