Autographa californica nuclear polyhedrosis virus DNA polymerase: measurements of processivity and strand displacement
- PMID: 10233952
- PMCID: PMC112534
- DOI: 10.1128/JVI.73.6.4908-4918.1999
Autographa californica nuclear polyhedrosis virus DNA polymerase: measurements of processivity and strand displacement
Abstract
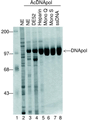
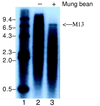
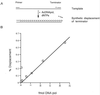
The DNA polymerase (DNApol) of Autographa californica nuclear polyhedrosis virus was purified to homogeneity from recombinant baculovirus-infected cells. DNApol was active in polymerase assays on singly primed M13 template, and full-length replicative form II product was synthesized at equimolar ratios of enzyme to template. The purified recombinant DNApol was shown to be processive by template challenge assay. Furthermore, DNApol was able to incorporate hundreds of nucleotides on an oligo(dT)-primed poly(dA) template with limiting amounts of polymerase. DNApol has moderate strand displacement activity, as it was active on nicked and gapped templates, and displaced a primer in a replication-dependent manner. Addition of saturating amounts of LEF-3, the viral single-stranded DNA-binding protein (SSB), increased the innate strand displacement ability of DNApol. However, when LEF-3 was added prior to the polymerase, it failed to stimulate DNApol replication on a singly primed M13 template because the helix-destabilizing activity of LEF-3 caused the primer to dissociate from the template. Escherichia coli SSB efficiently substituted for LEF-3 in the replication of a nicked template, suggesting that specific protein-protein interactions were not required for strand displacement in this assay.
Figures






Similar articles
-
Autographa californica multiple nucleopolyhedrovirus DNA polymerase C terminus is required for nuclear localization and viral DNA replication.J Virol. 2014 Sep;88(18):10918-33. doi: 10.1128/JVI.01167-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008932 Free PMC article.
-
A betabaculovirus DNA polymerase cannot substitute for the DNA polymerase of the alphabaculovirus Autographa californica nucleopolyhedrovirus.Arch Virol. 2017 Nov;162(11):3487-3492. doi: 10.1007/s00705-017-3468-0. Epub 2017 Jul 20. Arch Virol. 2017. PMID: 28730520
-
Expression, purification and characterization of the Spodoptera littoralis nucleopolyhedrovirus (SpliNPV) DNA polymerase and interaction with the SpliNPV non-hr origin of DNA replication.J Gen Virol. 2001 Jul;82(Pt 7):1767-1776. doi: 10.1099/0022-1317-82-7-1767. J Gen Virol. 2001. PMID: 11413389
-
The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein.J Virol. 1995 Jun;69(6):3924-8. doi: 10.1128/JVI.69.6.3924-3928.1995. J Virol. 1995. PMID: 7745748 Free PMC article.
-
The deficiency in nuclear localization signal of Neodiprion lecontei nucleopolyhedrovirus DNA polymerase prevents rescue of viral DNA replication and virus production in dnapol-null Autographa californica multiple nucleopolyhedrovirus.Virus Res. 2019 Jun;266:52-57. doi: 10.1016/j.virusres.2019.04.005. Epub 2019 Apr 13. Virus Res. 2019. PMID: 30991090
Cited by
-
Baculovirus Genetic Diversity and Population Structure.Viruses. 2025 Jan 22;17(2):142. doi: 10.3390/v17020142. Viruses. 2025. PMID: 40006898 Free PMC article. Review.
-
Genetic requirements for homologous recombination in Autographa californica nucleopolyhedrovirus.J Virol. 2002 Sep;76(18):9323-34. doi: 10.1128/jvi.76.18.9323-9334.2002. J Virol. 2002. PMID: 12186915 Free PMC article.
-
Baculovirus replication factor LEF-1 is a DNA primase.J Virol. 2002 Mar;76(5):2287-97. doi: 10.1128/jvi.76.5.2287-2297.2002. J Virol. 2002. PMID: 11836407 Free PMC article.
-
Autographa californica multiple nucleopolyhedrovirus DNA polymerase C terminus is required for nuclear localization and viral DNA replication.J Virol. 2014 Sep;88(18):10918-33. doi: 10.1128/JVI.01167-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008932 Free PMC article.
-
Generation of Variability in Chrysodeixis includens Nucleopolyhedrovirus (ChinNPV): The Role of a Single Variant.Viruses. 2021 Sep 22;13(10):1895. doi: 10.3390/v13101895. Viruses. 2021. PMID: 34696324 Free PMC article.
References
-
- Barrett J W, Lauzon H A M, Mercuri P S, Krell P J, Sohi S S, Arif B M. The putative LEF-1 proteins from two distinct Choristuneura fumiferana multiple nucleopolyhedroviruses share domain homology to eukaryotic primases. Virus Genes. 1996;13:229. - PubMed
-
- Blanco L, Bernad A, Lazaro J M, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage φ29 DNA polymerase. J Biol Chem. 1989;264:8935–8940. - PubMed
-
- Field J, Gronostajski R M, Hurwitz J. Properties of the adenovirus DNA polymerase. J Biol Chem. 1984;259:9487–9495. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

