Microglial activation varies in different models of Creutzfeldt-Jakob disease
- PMID: 10233972
- PMCID: PMC112554
- DOI: 10.1128/JVI.73.6.5089-5097.1999
Microglial activation varies in different models of Creutzfeldt-Jakob disease
Abstract
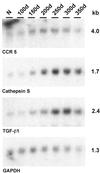
Progressive changes in host mRNA expression can illuminate crucial pathogenetic pathways in infectious disease. We examined general and specific approaches to mRNA expression in three rodent models of Creutzfeldt-Jakob disease (CJD). Each of these models displays distinctive neuropathology. Although mRNAs for the chemokine receptor CCR5, the lysosomal protease cathepsin S, and the pleiotropic cytokine transforming growth factor beta1 (TGF-beta1) were progressively upregulated in rodent CJD, the temporal patterns and peak magnitudes of each of these transcripts varied substantially among models. Cathepsin S and TGF-beta1 were elevated more than 15-fold in mice and rats infected with two different CJD strains, but not in CJD-infected hamsters. In rats, an early activation of microglial transcripts preceded obvious deposits of prion protein (PrP) amyloid. However, in each of the three CJD models, the upregulation of CCR5, cathepsin S, and TGF-beta1 was variable with respect to the onset of PrP pathology. These results show glial cell involvement varies as a consequence of the agent strain and species infected. Although neurons are generally assumed to be the primary sites for agent replication and abnormal PrP formation, microglia may be targeted by some agent strains. In such instances, microglia can both process PrP to become amyloid and can enhance neuronal destruction. Because microglia can participate in agent clearance, they may also act as chronic reservoirs of infectivity. Finally, the results here strongly suggest that TGF-beta1 can be an essential signal for amyloid deposition.
Figures




References
-
- Aguzzi A, Weissmann C. Prion research: the next frontiers. Nature. 1997;389:795–798. - PubMed
-
- Akowitz A, Sklaviadis T, Manuelidis E E, Manuelidis L. Nuclease-resistant polyadenylated RNAs of significant size are detected by PCR in highly purified Creutzfeldt-Jakob disease preparations. Microb Pathog. 1990;9:33–45. - PubMed
-
- Berglund E O, Ranscht B. Molecular cloning and in situ localization of the human contactin gene (CNTN1) on chromosome 12q11-q12. Genomics. 1994;21:571–582. - PubMed
-
- Bruce M E, McConnell I, Fraser H, Dickinson A G. The disease characteristics of different strains of scrapie in Sinc congenic mouse lines: implications for the nature of the agent and host control of pathogenesis. J Gen Virol. 1991;72:595–603. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

