Assessment of the allosteric mechanism of aspartate transcarbamoylase based on the crystalline structure of the unregulated catalytic subunit
- PMID: 10318893
- PMCID: PMC21869
- DOI: 10.1073/pnas.96.10.5388
Assessment of the allosteric mechanism of aspartate transcarbamoylase based on the crystalline structure of the unregulated catalytic subunit
Abstract
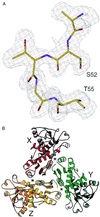
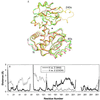
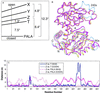
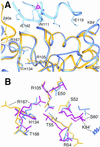
The lack of knowledge of the three-dimensional structure of the trimeric, catalytic (C) subunit of aspartate transcarbamoylase (ATCase) has impeded understanding of the allosteric regulation of this enzyme and left unresolved the mechanism by which the active, unregulated C trimers are inactivated on incorporation into the unliganded (taut or T state) holoenzyme. Surprisingly, the isolated C trimer, based on the 1.9-A crystal structure reported here, resembles more closely the trimers in the T state enzyme than in the holoenzyme:bisubstrate-analog complex, which has been considered as the active, relaxed (R) state enzyme. Unlike the C trimer in either the T state or bisubstrate-analog-bound holoenzyme, the isolated C trimer lacks 3-fold symmetry, and the active sites are partially disordered. The flexibility of the C trimer, contrasted to the highly constrained T state ATCase, suggests that regulation of the holoenzyme involves modulating the potential for conformational changes essential for catalysis. Large differences in structure between the active C trimer and the holoenzyme:bisubstrate-analog complex call into question the view that this complex represents the activated R state of ATCase.
Figures
Similar articles
-
Binding of bisubstrate analog promotes large structural changes in the unregulated catalytic trimer of aspartate transcarbamoylase: implications for allosteric regulation.Proc Natl Acad Sci U S A. 2000 May 9;97(10):5077-82. doi: 10.1073/pnas.090087197. Proc Natl Acad Sci U S A. 2000. PMID: 10805770 Free PMC article.
-
Peptide-protein interaction markedly alters the functional properties of the catalytic subunit of aspartate transcarbamoylase.Protein Sci. 1993 Jan;2(1):103-12. doi: 10.1002/pro.5560020111. Protein Sci. 1993. PMID: 8443583 Free PMC article.
-
Charge neutralization in the active site of the catalytic trimer of aspartate transcarbamoylase promotes diverse structural changes.Protein Sci. 2017 Nov;26(11):2221-2228. doi: 10.1002/pro.3277. Epub 2017 Sep 30. Protein Sci. 2017. PMID: 28833948 Free PMC article.
-
Structure and mechanisms of Escherichia coli aspartate transcarbamoylase.Acc Chem Res. 2012 Mar 20;45(3):444-53. doi: 10.1021/ar200166p. Epub 2011 Oct 19. Acc Chem Res. 2012. PMID: 22011033 Free PMC article. Review.
-
Allosteric regulation of catalytic activity: Escherichia coli aspartate transcarbamoylase versus yeast chorismate mutase.Microbiol Mol Biol Rev. 2001 Sep;65(3):404-21, table of contents. doi: 10.1128/MMBR.65.3.404-421.2001. Microbiol Mol Biol Rev. 2001. PMID: 11528003 Free PMC article. Review.
Cited by
-
Crystallographic snapshots of the complete catalytic cycle of the unregulated aspartate transcarbamoylase from Bacillus subtilis.J Mol Biol. 2011 Aug 5;411(1):190-200. doi: 10.1016/j.jmb.2011.05.036. Epub 2011 May 31. J Mol Biol. 2011. PMID: 21663747 Free PMC article.
-
Allostery and cooperativity in Escherichia coli aspartate transcarbamoylase.Arch Biochem Biophys. 2012 Mar 15;519(2):81-90. doi: 10.1016/j.abb.2011.10.024. Epub 2011 Dec 16. Arch Biochem Biophys. 2012. PMID: 22198283 Free PMC article. Review.
-
Dihydroorotase from the hyperthermophile Aquifex aeolicus is activated by stoichiometric association with aspartate transcarbamoylase and forms a one-pot reactor for pyrimidine biosynthesis.Biochemistry. 2009 Feb 3;48(4):766-78. doi: 10.1021/bi801831r. Biochemistry. 2009. PMID: 19128030 Free PMC article.
-
Quaternary structure of hemoglobin in solution.Proc Natl Acad Sci U S A. 2003 Jan 21;100(2):517-20. doi: 10.1073/pnas.232715799. Epub 2003 Jan 13. Proc Natl Acad Sci U S A. 2003. PMID: 12525687 Free PMC article.
-
ELISA: structure-function inferences based on statistically significant and evolutionarily inspired observations.BMC Bioinformatics. 2003 Sep 2;4:34. doi: 10.1186/1471-2105-4-34. Epub 2003 Sep 2. BMC Bioinformatics. 2003. PMID: 12952559 Free PMC article.
References
-
- Monod J, Wyman J, Changeux J-P. J Mol Biol. 1965;12:88–118. - PubMed
-
- Howlett G J, Blackburn M N, Compton J G, Schachman H K. Biochemistry. 1977;16:5091–5099. - PubMed
-
- Schachman H K. J Biol Chem. 1988;263:18583–18586. - PubMed
-
- Eisenstein E, Markby D W, Schachman H K. Biochemistry. 1990;29:3724–3731. - PubMed
-
- Collins K D, Stark G R. J Biol Chem. 1971;246:6599–6605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

