Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays
- PMID: 10318894
- PMCID: PMC21870
- DOI: 10.1073/pnas.96.10.5394
Clonal selection and in vivo quantitation of protein interactions with protein-fragment complementation assays
Erratum in
- Proc Natl Acad Sci U S A 1999 Jun 22;96(13):7610
Abstract
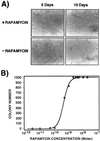
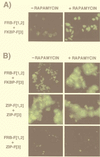
Two strategies are described for detecting constitutive or induced protein-protein interactions in intact mammalian cells; these strategies are based on oligomerization domain-assisted complementation of rationally designed fragments of the murine enzyme dihydrofolate reductase (DHFR; EC 1.5.1.3). We describe a dominant clonal-selection assay of stably transfected cells expressing partner proteins FKBP (FK506 binding protein) and FRAP (FKBP-rapamycin binding protein) fused to DHFR fragments and show a rapamycin dose-dependent survival of clones that requires approximately 25 molecules of reconstituted DHFR per cell. A fluorescence assay also is described, based on stoichiometric binding of fluorescein-methotrexate to reconstituted DHFR in vivo. Formation of the FKBP-rapamycin-FRAP complex is detected in stably and transiently transfected cells. Quantitative rapamycin dose-dependence of this complex is shown to be consistent with in vitro binding and distinguishable from a known constitutive interaction of FKBP and FRAP. We also show that this strategy can be applied to study membrane protein receptors, demonstrating dose-dependent activation of the erythropoietin receptor by ligands. The combination of these clonal-selection and fluorescence assays in intact mammalian cells makes possible selection by simple survival, flow cytometry, or both. High-throughput drug screening and quantitative analysis of induction or disruption of protein-protein interactions are also made possible.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

